Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions
- PMID: 28977401
- PMCID: PMC5737381
- DOI: 10.1093/nar/gkx699
Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions
Abstract
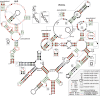
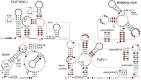
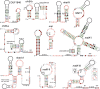
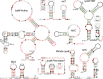
The discovery of structured non-coding RNAs (ncRNAs) in bacteria can reveal new facets of biology and biochemistry. Comparative genomics analyses executed by powerful computer algorithms have successfully been used to uncover many novel bacterial ncRNA classes in recent years. However, this general search strategy favors the discovery of more common ncRNA classes, whereas progressively rarer classes are correspondingly more difficult to identify. In the current study, we confront this problem by devising several methods to select subsets of intergenic regions that can concentrate these rare RNA classes, thereby increasing the probability that comparative sequence analysis approaches will reveal their existence. By implementing these methods, we discovered 224 novel ncRNA classes, which include ROOL RNA, an RNA class averaging 581 nt and present in multiple phyla, several highly conserved and widespread ncRNA classes with properties that suggest sophisticated biochemical functions and a multitude of putative cis-regulatory RNA classes involved in a variety of biological processes. We expect that further research on these newly found RNA classes will reveal additional aspects of novel biology, and allow for greater insights into the biochemistry performed by ncRNAs.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- Meyer M.M. The role of mRNA structure in bacterial translational regulation. Wiley Interdiscip. Rev. RNA. 2017; 8:e1370. - PubMed
-
- Barquist L., Vogel J.. Accelerating discovery and functional analysis of small RNAs with new technologies. Annu. Rev. Genet. 2015; 49:367–394. - PubMed
-
- Wachter A. Gene regulation by structured mRNA elements. Trends Genet. 2014; 30:172–181. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

