A pathway for low zinc homeostasis that is conserved in animals and acts in parallel to the pathway for high zinc homeostasis
- PMID: 28977437
- PMCID: PMC5714235
- DOI: 10.1093/nar/gkx762
A pathway for low zinc homeostasis that is conserved in animals and acts in parallel to the pathway for high zinc homeostasis
Abstract
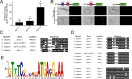
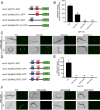
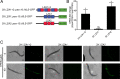
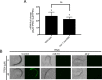
The essential element zinc plays critical roles in biology. High zinc homeostasis mechanisms are beginning to be defined in animals, but low zinc homeostasis is poorly characterized. We investigated low zinc homeostasis in Caenorhabditis elegans because the genome encodes 14 evolutionarily conserved Zrt, Irt-like protein (ZIP) zinc transporter family members. Three C. elegans zipt genes were regulated in zinc-deficient conditions; these promoters contained an evolutionarily conserved motif that we named the low zinc activation (LZA) element that was both necessary and sufficient for activation of transcription in response to zinc deficiency. These results demonstrated that the LZA element is a critical part of the low zinc homeostasis pathway. Transcriptional regulation of the LZA element required the transcription factor ELT-2 and mediator complex member MDT-15. We investigated conservation in mammals by analyzing LZA element function in human cultured cells; the LZA element-mediated transcriptional activation in response to zinc deficiency in cells, suggesting a conserved pathway of low zinc homeostasis. We propose that the pathway for low zinc homeostasis, which includes the LZA element and ZIP transporters, acts in parallel to the pathway for high zinc homeostasis, which includes the HZA element, HIZR-1 transcription factor and cation diffusion facilitator transporters.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Gaither L.A., Eide D.J.. Eukaryotic zinc transporters and their regulation. Biometals. 2001; 14:251–270. - PubMed
-
- Vallee B.L., Falchuk K.H.. The biochemical basis of zinc physiology. Physiol. Rev. 1993; 73:79–118. - PubMed
-
- Andreini C., Banci L., Ivano B., Rosato A.. Counting the zinc-proteins encoded in the human genome. J. Proteome Res. 2005; 5:196–201. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

