The genomic landscape of African populations in health and disease
- PMID: 28977439
- PMCID: PMC6075021
- DOI: 10.1093/hmg/ddx253
The genomic landscape of African populations in health and disease
Abstract
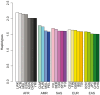
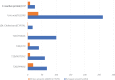
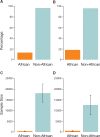
A deeper appreciation of the complex architecture of African genomes is critical to the global effort to understand human history, biology and differential distribution of disease by geography and ancestry. Here, we report on how the growing engagement of African populations in genome science is providing new insights into the forces that shaped human genomes before and after the Out-of-Africa migrations. As a result of this human evolutionary history, African ancestry populations have the greatest genomic diversity in the world, and this diversity has important ramifications for genomic research. In the case of pharmacogenomics, for instance, variants of consequence are not limited to those identified in other populations, and diversity within African ancestry populations precludes summarizing risk across different African ethnic groups. Exposure of Africans to fatal pathogens, such as Plasmodium falciparum, Lassa Virus and Trypanosoma brucei rhodesiense, has resulted in elevated frequencies of alleles conferring survival advantages for infectious diseases, but that are maladaptive in modern-day environments. Illustrating with cardiometabolic traits, we show that while genomic research in African ancestry populations is still in early stages, there are already many examples of novel and African ancestry-specific disease loci that have been discovered. Furthermore, the shorter haplotypes in African genomes have facilitated fine-mapping of loci discovered in other human ancestry populations. Given the insights already gained from the interrogation of African genomes, it is imperative to continue and increase our efforts to describe genomic risk in and across African ancestry populations.
Published by Oxford University Press 2017. This work is written by US Government employees and is in the public domain in the US.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

