Plant organellar DNA primase-helicase synthesizes RNA primers for organellar DNA polymerases using a unique recognition sequence
- PMID: 28977480
- PMCID: PMC5737085
- DOI: 10.1093/nar/gkx745
Plant organellar DNA primase-helicase synthesizes RNA primers for organellar DNA polymerases using a unique recognition sequence
Abstract
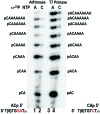
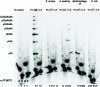
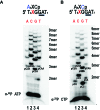
DNA primases recognize single-stranded DNA (ssDNA) sequences to synthesize RNA primers during lagging-strand replication. Arabidopsis thaliana encodes an ortholog of the DNA primase-helicase from bacteriophage T7, dubbed AtTwinkle, that localizes in chloroplasts and mitochondria. Herein, we report that AtTwinkle synthesizes RNA primers from a 5'-(G/C)GGA-3' template sequence. Within this sequence, the underlined nucleotides are cryptic, meaning that they are essential for template recognition but are not instructional during RNA synthesis. Thus, in contrast to all primases characterized to date, the sequence recognized by AtTwinkle requires two nucleotides (5'-GA-3') as a cryptic element. The divergent zinc finger binding domain (ZBD) of the primase module of AtTwinkle may be responsible for template sequence recognition. During oligoribonucleotide synthesis, AtTwinkle shows a strong preference for rCTP as its initial ribonucleotide and a moderate preference for rGMP or rCMP incorporation during elongation. RNA products synthetized by AtTwinkle are efficiently used as primers for plant organellar DNA polymerases. In sum, our data strongly suggest that AtTwinkle primes organellar DNA polymerases during lagging strand synthesis in plant mitochondria and chloroplast following a primase-mediated mechanism. This mechanism contrasts to lagging-strand DNA replication in metazoan mitochondria, in which transcripts synthesized by mitochondrial RNA polymerase prime mitochondrial DNA polymerase γ.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

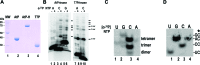




Similar articles
-
The plant organellar primase-helicase directs template recognition and primosome assembly via its zinc finger domain.BMC Plant Biol. 2023 Oct 6;23(1):467. doi: 10.1186/s12870-023-04477-4. BMC Plant Biol. 2023. PMID: 37803262 Free PMC article.
-
A Bacteriophage-Derived Primase-Helicase Orchestrates Plant Organellar DNA Replication.Physiol Plant. 2025 Jul-Aug;177(4):e70379. doi: 10.1111/ppl.70379. Physiol Plant. 2025. PMID: 40620047 Free PMC article.
-
Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro.Proc Natl Acad Sci U S A. 2008 Aug 12;105(32):11122-7. doi: 10.1073/pnas.0805399105. Epub 2008 Aug 6. Proc Natl Acad Sci U S A. 2008. PMID: 18685103 Free PMC article.
-
Initiating DNA replication: a matter of prime importance.Biochem Soc Trans. 2019 Feb 28;47(1):351-356. doi: 10.1042/BST20180627. Epub 2019 Jan 15. Biochem Soc Trans. 2019. PMID: 30647143 Free PMC article. Review.
-
Choreography of bacteriophage T7 DNA replication.Curr Opin Chem Biol. 2011 Oct;15(5):580-6. doi: 10.1016/j.cbpa.2011.07.024. Epub 2011 Sep 9. Curr Opin Chem Biol. 2011. PMID: 21907611 Free PMC article. Review.
Cited by
-
Plant Organelle Genome Replication.Plants (Basel). 2019 Sep 21;8(10):358. doi: 10.3390/plants8100358. Plants (Basel). 2019. PMID: 31546578 Free PMC article. Review.
-
Plant organellar DNA polymerases are replicative and translesion DNA synthesis polymerases.Nucleic Acids Res. 2017 Oct 13;45(18):10751-10763. doi: 10.1093/nar/gkx744. Nucleic Acids Res. 2017. PMID: 28977655 Free PMC article.
-
The N-terminal domain of human mitochondrial helicase Twinkle has DNA-binding activity crucial for supporting processive DNA synthesis by polymerase γ.J Biol Chem. 2023 Jan;299(1):102797. doi: 10.1016/j.jbc.2022.102797. Epub 2022 Dec 14. J Biol Chem. 2023. PMID: 36528058 Free PMC article.
-
Plant organellar DNA polymerases repair double-stranded breaks by microhomology-mediated end-joining.Nucleic Acids Res. 2019 Apr 8;47(6):3028-3044. doi: 10.1093/nar/gkz039. Nucleic Acids Res. 2019. PMID: 30698803 Free PMC article.
-
The Dictyostelium discoideum homologue of Twinkle, Twm1, is a mitochondrial DNA helicase, an active primase and promotes mitochondrial DNA replication.BMC Mol Biol. 2018 Dec 19;19(1):12. doi: 10.1186/s12867-018-0114-7. BMC Mol Biol. 2018. PMID: 30563453 Free PMC article.
References
-
- Shutt T.E., Gray M.W.. Bacteriophage origins of mitochondrial replication and transcription proteins. Trends Genet. 2006; 22:90–95. - PubMed
-
- Shutt T.E., Gray M.W.. Twinkle, the mitochondrial replicative DNA helicase, is widespread in the eukaryotic radiation and may also be the mitochondrial DNA primase in most eukaryotes. J. Mol. Evol. 2006; 62:588–599. - PubMed
-
- Masters B.S., Stohl L.L., Clayton D.A.. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell. 1987; 51:89–99. - PubMed
-
- Hamdan S.M., Richardson C.C.. Motors, switches, and contacts in the replisome. Annu. Rev. Biochem. 2009; 78:205–243. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

