DNA residence time is a regulatory factor of transcription repression
- PMID: 28977492
- PMCID: PMC5737411
- DOI: 10.1093/nar/gkx728
DNA residence time is a regulatory factor of transcription repression
Abstract
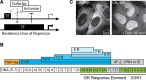
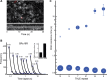
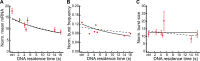
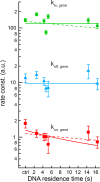
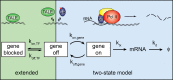
Transcription comprises a highly regulated sequence of intrinsically stochastic processes, resulting in bursts of transcription intermitted by quiescence. In transcription activation or repression, a transcription factor binds dynamically to DNA, with a residence time unique to each factor. Whether the DNA residence time is important in the transcription process is unclear. Here, we designed a series of transcription repressors differing in their DNA residence time by utilizing the modular DNA binding domain of transcription activator-like effectors (TALEs) and varying the number of nucleotide-recognizing repeat domains. We characterized the DNA residence times of our repressors in living cells using single molecule tracking. The residence times depended non-linearly on the number of repeat domains and differed by more than a factor of six. The factors provoked a residence time-dependent decrease in transcript level of the glucocorticoid receptor-activated gene SGK1. Down regulation of transcription was due to a lower burst frequency in the presence of long binding repressors and is in accordance with a model of competitive inhibition of endogenous activator binding. Our single molecule experiments reveal transcription factor DNA residence time as a regulatory factor controlling transcription repression and establish TALE-DNA binding domains as tools for the temporal dissection of transcription regulation.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Levine M., Manley J.L.. Transcriptional repression of eukaryotic promoters. Cell. 1989; 59:405–408. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

