N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3'-end processing
- PMID: 28977517
- PMCID: PMC5737695
- DOI: 10.1093/nar/gkx778
N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3'-end processing
Abstract
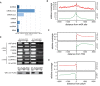
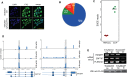
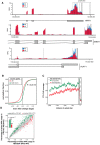
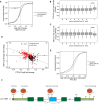
N6-methyladenosine (m6A) is the most abundant base modification found in messenger RNAs (mRNAs). The discovery of FTO as the first m6A mRNA demethylase established the concept of reversible RNA modification. Here, we present a comprehensive transcriptome-wide analysis of RNA demethylation and uncover FTO as a potent regulator of nuclear mRNA processing events such as alternative splicing and 3΄ end mRNA processing. We show that FTO binds preferentially to pre-mRNAs in intronic regions, in the proximity of alternatively spliced (AS) exons and poly(A) sites. FTO knockout (KO) results in substantial changes in pre-mRNA splicing with prevalence of exon skipping events. The alternative splicing effects of FTO KO anti-correlate with METTL3 knockdown suggesting the involvement of m6A. Besides, deletion of intronic region that contains m6A-linked DRACH motifs partially rescues the FTO KO phenotype in a reporter system. All together, we demonstrate that the splicing effects of FTO are dependent on the catalytic activity in vivo and are mediated by m6A. Our results reveal for the first time the dynamic connection between FTO RNA binding and demethylation activity that influences several mRNA processing events.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012; 485:201–206. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

