CRISPR-mediated isolation of specific megabase segments of genomic DNA
- PMID: 28977642
- PMCID: PMC5737698
- DOI: 10.1093/nar/gkx749
CRISPR-mediated isolation of specific megabase segments of genomic DNA
Abstract
Megabase-sized, complex, repetitive regions of genomes are poorly studied, due to the technical and computational challenges inherent to both assembling precise reference sequences and accurately assessing structural variation across contiguous megabase DNA regions. Here we describe a strategy to overcome these challenges, CISMR (CRISPR-mediated isolation of specific megabase-sized regions of the genome), which enables us to perform targeted isolation of contiguous megabase-sized segments of the genome. Direct sequencing of the purified DNA segments can have >100-fold enrichment of the target region, thus enabling the exploration of both DNA sequence and structural diversity of complex genomic regions in any species.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
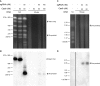

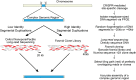
Similar articles
-
Enrichment of megabase-sized DNA molecules for single-molecule optical mapping and next-generation sequencing.Sci Rep. 2017 Dec 20;7(1):17893. doi: 10.1038/s41598-017-18091-6. Sci Rep. 2017. PMID: 29263336 Free PMC article.
-
μLAS technology for DNA isolation coupled to Cas9-assisted targeting for sequencing and assembly of a 30 kb region in plant genome.Nucleic Acids Res. 2019 Sep 5;47(15):8050-8060. doi: 10.1093/nar/gkz632. Nucleic Acids Res. 2019. PMID: 31505675 Free PMC article.
-
Electrophoretic karyotype analysis. Pulsed field gel electrophoresis.Methods Mol Biol. 1994;29:221-52. doi: 10.1385/0-89603-289-2:221. Methods Mol Biol. 1994. PMID: 8032409 No abstract available.
-
[DNA Fragment Enrichment for High-Throughput Sequencing].Mol Biol (Mosk). 2023 May-Jun;57(3):440-457. Mol Biol (Mosk). 2023. PMID: 37326047 Review. Russian.
-
Chromosomal localization of genes through pulsed-field gel electrophoresis techniques.Methods Mol Biol. 1996;53:79-88. doi: 10.1385/0-89603-319-8:79. Methods Mol Biol. 1996. PMID: 8925008 Review. No abstract available.
Cited by
-
Repetitive DNA Sequences in the Human Y Chromosome and Male Infertility.Front Cell Dev Biol. 2022 Jul 13;10:831338. doi: 10.3389/fcell.2022.831338. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35912115 Free PMC article. Review.
-
How Helpful May Be a CRISPR/Cas-Based System for Food Traceability?Foods. 2024 Oct 25;13(21):3397. doi: 10.3390/foods13213397. Foods. 2024. PMID: 39517184 Free PMC article. Review.
-
Optimising Guide RNA Production for Multiplexed Cas9-Targeted Nanopore Sequencing to Detect Pathogens.Mol Biotechnol. 2025 Sep 7. doi: 10.1007/s12033-025-01510-9. Online ahead of print. Mol Biotechnol. 2025. PMID: 40914913
-
Not Only Editing: A Cas-Cade of CRISPR/Cas-Based Tools for Functional Genomics in Plants and Animals.Int J Mol Sci. 2024 Mar 13;25(6):3271. doi: 10.3390/ijms25063271. Int J Mol Sci. 2024. PMID: 38542245 Free PMC article. Review.
-
Leveraging the power of long reads for targeted sequencing.Genome Res. 2024 Nov 20;34(11):1701-1718. doi: 10.1101/gr.279168.124. Genome Res. 2024. PMID: 39567237 Free PMC article. Review.
References
-
- Watson C.T., Steinberg K.M., Huddleston J., Warren R.L., Malig M., Schein J., Willsey A.J., Joy J.B., Scott J.K., Graves T.A. et al. . Complete haplotype sequence of the human immunoglobulin heavy-chain variable, diversity, and joining genes and characterization of allelic and copy-number variation. Am. J. Hum. Genet. 2013; 92:530–546. - PMC - PubMed
-
- Skaletsky H., Kuroda-Kawaguchi T., Minx P.J., Cordum H.S., Hillier L., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T. et al. . The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003; 423:825–837. - PubMed
-
- Hughes J.F., Rozen S.. Genomics and genetics of human and primate y chromosomes. Annu. Rev. Genomics Hum. Genet. 2012; 13:83–108. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

