VDJdb: a curated database of T-cell receptor sequences with known antigen specificity
- PMID: 28977646
- PMCID: PMC5753233
- DOI: 10.1093/nar/gkx760
VDJdb: a curated database of T-cell receptor sequences with known antigen specificity
Abstract
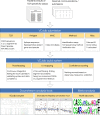
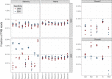
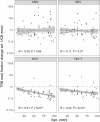
The ability to decode antigen specificities encapsulated in the sequences of rearranged T-cell receptor (TCR) genes is critical for our understanding of the adaptive immune system and promises significant advances in the field of translational medicine. Recent developments in high-throughput sequencing methods (immune repertoire sequencing technology, or RepSeq) and single-cell RNA sequencing technology have allowed us to obtain huge numbers of TCR sequences from donor samples and link them to T-cell phenotypes. However, our ability to annotate these TCR sequences still lags behind, owing to the enormous diversity of the TCR repertoire and the scarcity of available data on T-cell specificities. In this paper, we present VDJdb, a database that stores and aggregates the results of published T-cell specificity assays and provides a universal platform that couples antigen specificities with TCR sequences. We demonstrate that VDJdb is a versatile instrument for the annotation of TCR repertoire data, enabling a concatenated view of antigen-specific TCR sequence motifs. VDJdb can be accessed at https://vdjdb.cdr3.net and https://github.com/antigenomics/vdjdb-db.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Bacher P., Scheffold A.. Flow-cytometric analysis of rare antigen-specific T cells. Cytometry A. 2013; 83:692–701. - PubMed
-
- Tickotsky N., Sagiv T., Prilusky J., Shifrut E., Friedman N.. McPAS-TCR: a manually-curated catalogue of pathology-associated T-cell receptor sequences. Bioinformatics. 2017; doi:10.1093/bioinformatics/btx286. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

