A 'new lease of life': FnCpf1 possesses DNA cleavage activity for genome editing in human cells
- PMID: 28977650
- PMCID: PMC5737432
- DOI: 10.1093/nar/gkx783
A 'new lease of life': FnCpf1 possesses DNA cleavage activity for genome editing in human cells
Abstract
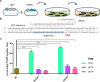
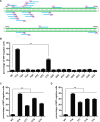
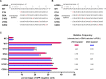
Cpf1 nucleases were recently reported to be highly specific and programmable nucleases with efficiencies comparable to those of SpCas9. AsCpf1 and LbCpf1 require a single crRNA and recognize a 5'-TTTN-3' protospacer adjacent motif (PAM) at the 5' end of the protospacer for genome editing. For widespread application in precision site-specific human genome editing, the range of sequences that AsCpf1 and LbCpf1 can recognize is limited due to the size of this PAM. To address this limitation, we sought to identify a novel Cpf1 nuclease with simpler PAM requirements. Specifically, here we sought to test and engineer FnCpf1, one reported Cpf1 nuclease (FnCpf1) only requires 5'-TTN-3' as a PAM but does not exhibit detectable levels of nuclease-induced indels at certain locus in human cells. Surprisingly, we found that FnCpf1 possesses DNA cleavage activity in human cells at multiple loci. We also comprehensively and quantitatively examined various FnCpf1 parameters in human cells, including spacer sequence, direct repeat sequence and the PAM sequence. Our study identifies FnCpf1 as a new member of the Cpf1 family for human genome editing with distinctive characteristics, which shows promise as a genome editing tool with the potential for both research and therapeutic applications.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
The Conspicuity of CRISPR-Cpf1 System as a Significant Breakthrough in Genome Editing.Curr Microbiol. 2018 Jan;75(1):107-115. doi: 10.1007/s00284-017-1406-8. Epub 2017 Nov 30. Curr Microbiol. 2018. PMID: 29189942 Review.
-
Engineering the Direct Repeat Sequence of crRNA for Optimization of FnCpf1-Mediated Genome Editing in Human Cells.Mol Ther. 2018 Nov 7;26(11):2650-2657. doi: 10.1016/j.ymthe.2018.08.021. Epub 2018 Sep 1. Mol Ther. 2018. PMID: 30274789 Free PMC article.
-
Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells.Nat Biotechnol. 2016 Aug;34(8):863-8. doi: 10.1038/nbt.3609. Epub 2016 Jun 6. Nat Biotechnol. 2016. PMID: 27272384
-
Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1.Mol Cell. 2017 Aug 17;67(4):633-645.e3. doi: 10.1016/j.molcel.2017.06.035. Epub 2017 Aug 3. Mol Cell. 2017. PMID: 28781234 Free PMC article.
-
CRISPR-Cpf1-mediated genome editing and gene regulation in human cells.Biotechnol Adv. 2019 Jan-Feb;37(1):21-27. doi: 10.1016/j.biotechadv.2018.10.013. Epub 2018 Nov 3. Biotechnol Adv. 2019. PMID: 30399413 Review.
Cited by
-
A resurrected ancestor of Cas12a expands target access and substrate recognition for nucleic acid editing and detection.Nat Biotechnol. 2024 Oct 31. doi: 10.1038/s41587-024-02461-3. Online ahead of print. Nat Biotechnol. 2024. PMID: 39482449
-
The Rise of the CRISPR/Cpf1 System for Efficient Genome Editing in Plants.Front Plant Sci. 2020 Mar 31;11:264. doi: 10.3389/fpls.2020.00264. eCollection 2020. Front Plant Sci. 2020. PMID: 32296449 Free PMC article. Review.
-
Nonspecific interactions between Cas12a and dsDNA located downstream of the PAM mediate target search and assist AsCas12a for DNA cleavage.Chem Sci. 2023 Mar 10;14(14):3839-3851. doi: 10.1039/d2sc05463a. eCollection 2023 Apr 5. Chem Sci. 2023. PMID: 37035707 Free PMC article.
-
The Conspicuity of CRISPR-Cpf1 System as a Significant Breakthrough in Genome Editing.Curr Microbiol. 2018 Jan;75(1):107-115. doi: 10.1007/s00284-017-1406-8. Epub 2017 Nov 30. Curr Microbiol. 2018. PMID: 29189942 Review.
-
PlmCas12e Utilizes Glu662 to Prevent Cleavage Site Occupation by Positively Charged Residues Before Target Strand Cleavage.Molecules. 2024 Oct 25;29(21):5036. doi: 10.3390/molecules29215036. Molecules. 2024. PMID: 39519677 Free PMC article.
References
-
- Liu N., Chen G.Q., Ning G.A., Shi H.B., Zhang C.L., Lu J.P., Mao L.J., Feng X.X., Liu X.H., Su Z.Z. et al. . Agrobacterium tumefaciens-mediated transformation: An efficient tool for insertional mutagenesis and targeted gene disruption in Harpophora oryzae. Microbiol. Res. 2016; 182:40–48. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

