A novel TPR-BEN domain interaction mediates PICH-BEND3 association
- PMID: 28977671
- PMCID: PMC5737856
- DOI: 10.1093/nar/gkx792
A novel TPR-BEN domain interaction mediates PICH-BEND3 association
Abstract
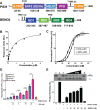
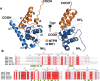
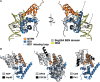
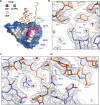
PICH is a DNA translocase required for the maintenance of chromosome stability in human cells. Recent data indicate that PICH co-operates with topoisomerase IIα to suppress pathological chromosome missegregation through promoting the resolution of ultra-fine anaphase bridges (UFBs). Here, we identify the BEN domain-containing protein 3 (BEND3) as an interaction partner of PICH in human cells in mitosis. We have purified full length PICH and BEND3 and shown that they exhibit a functional biochemical interaction in vitro. We demonstrate that the PICH-BEND3 interaction occurs via a novel interface between a TPR domain in PICH and a BEN domain in BEND3, and have determined the crystal structure of this TPR-BEN complex at 2.2 Å resolution. Based on the structure, we identified amino acids important for the TPR-BEN domain interaction, and for the functional interaction of the full-length proteins. Our data reveal a proposed new function for BEND3 in association with PICH, and the first example of a specific protein-protein interaction mediated by a BEN domain.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

