An unconventional family 1 uracil DNA glycosylase in Nitratifractor salsuginis
- PMID: 28977725
- PMCID: PMC5716868
- DOI: 10.1111/febs.14285
An unconventional family 1 uracil DNA glycosylase in Nitratifractor salsuginis
Abstract
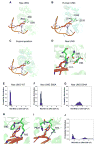
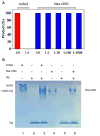
The uracil DNA glycosylase superfamily consists of at least six families with a diverse specificity toward DNA base damage. Family 1 uracil N-glycosylase (UNG) exhibits exclusive specificity on uracil-containing DNA. Here, we report a family 1 UNG homolog from Nitratifractor salsuginis with distinct biochemical features that differentiate it from conventional family 1 UNGs. Globally, the crystal structure of N. salsuginisUNG shows a few additional secondary structural elements. Biochemical and enzyme kinetic analysis, coupled with structural determination, molecular modeling, and molecular dynamics simulations, shows that N. salsuginisUNG contains a salt bridge network that plays an important role in DNA backbone interactions. Disruption of the amino acid residues involved in the salt bridges greatly impedes the enzymatic activity. A tyrosine residue in motif 1 (GQDPY) is one of the distinct sequence features setting family 1 UNG apart from other families. The crystal structure of Y81G mutant indicates that several subtle changes may account for its inactivity. Unlike the conventional family 1 UNG enzymes, N. salsuginisUNG is not inhibited by Ugi, a potent inhibitor specific for family 1 UNG. This study underscores the diversity of paths that a uracil DNA glycosylase may take to acquire its unique structural and biochemical properties during evolution.
Database: Structure data are available in the PDB under accession numbers 5X3G and 5X3H.
Keywords: DNA repair; deamination; protein-DNA interactions; salt bridge; uracil DNA glycosylase inhibitor.
© 2017 Federation of European Biochemical Societies.
Conflict of interest statement
Figures






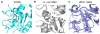

References
-
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–15. - PubMed
-
- Shapiro R. Damage to DNA caused by hydrolysis. In: Seeberg E, Kleppe K, editors. Chromosome Damage and Repair. Plenum Press; New York: 1981. pp. 3–18.
-
- Suzuki T, Yamaoka R, Nishi M, Ide H, Makino K. Isolation and characterization of a novel product, 2′-deoxyoxanosine, from 2′-deoxyguanosine, oligodeoxynucleotide and calf thymus DNA treated by nitrous-acid and nitric-oxide. J Am Chem Soc. 1996;118:2515–2516.
-
- Parikh SS, Putnam CD, Tainer JA. Lessons learned from structural results on uracil-DNA glycosylase. Mutat Res. 2000;460:183–99. - PubMed
-
- Krokan HE, Drablos F, Slupphaug G. Uracil in DNA--occurrence, consequences and repair. Oncogene. 2002;21:8935–48. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

