Gene expression profiling to predict recurrence of advanced squamous cell carcinoma of the tongue: discovery and external validation
- PMID: 28977904
- PMCID: PMC5617464
- DOI: 10.18632/oncotarget.18692
Gene expression profiling to predict recurrence of advanced squamous cell carcinoma of the tongue: discovery and external validation
Abstract
Objectives: To establish a prognostic signature for locally advanced tongue squamous cell carcinoma (TSCC) patients treated with surgery.
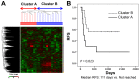
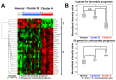
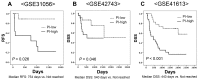
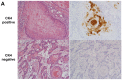
Results: In the discovery study, unsupervised hierarchical clustering analysis identified two clusters which differentiated the Kaplan-Meier curves of RFS [median RFS, 111 days vs. not reached; log-rank test, P = 0.023]. The 30 genes identified were combined into a dichotomous PI. In the validation cohort, classification according to the PI was associated with RFS [median RFS, 754 days vs. not reached; log-rank test, P = 0.026 in GSE31056] and DSS [median DSS, 540 days vs. not reached; log-rank test, P = 0.046 in GSE42743 and 443 days vs. not reached; P < 0.001 in GSE41613]. Among genes, positive immunohistochemical staining of cytokeratin 4 was associated with favorable prognostic values for RFS (hazard ratio (HR), 0.591, P = 0.045) and DSS (HR, 0.333, P = 0.004).
Materials and methods: We conducted gene expression profiling of 26 clinicopathologically homogeneous advanced TSCC tissue samples using cDNA microarray as a discovery study. Candidate genes were screened using clustering analysis and univariate Cox regression analysis for relapse-free survival (RFS). These were combined into a prognostic index (PI), which was validated using three public microarray datasets of tongue and oral cancer (123 patients). Some genes identified in discovery were immunohistochemically examined for protein expression in another 127 TSCC patients.
Conclusion: We identified robust molecular markers that showed significant associations with prognosis in TSCC patients. Gene expression profiling data were successfully converted to protein expression profiling data.
Keywords: cytokeratin 4; gene expression profile; keratin 4; oral squamous cell carcinoma; tongue squamous cell carcinoma.
Conflict of interest statement
CONFLICTS OF INTEREST No competing financial interests exist.
Figures

Similar articles
-
Overexpression of Hippo pathway effector TAZ in tongue squamous cell carcinoma: correlation with clinicopathological features and patients' prognosis.J Oral Pathol Med. 2013 Nov;42(10):747-54. doi: 10.1111/jop.12062. Epub 2013 Mar 29. J Oral Pathol Med. 2013. PMID: 23551691
-
A preliminary study of IgG4 expression and its prognostic significance in oral squamous cell carcinoma.BMC Cancer. 2024 Mar 4;24(1):294. doi: 10.1186/s12885-024-12048-5. BMC Cancer. 2024. PMID: 38438903 Free PMC article.
-
Identification of a novel immune gene panel in tongue squamous cell carcinoma.Am J Transl Res. 2022 May 15;14(5):2801-2824. eCollection 2022. Am J Transl Res. 2022. PMID: 35702068 Free PMC article.
-
Assessment of the prognostic and predictive utility of the Breast Cancer Index (BCI): an NCIC CTG MA.14 study.Breast Cancer Res. 2016 Jan 4;18(1):1. doi: 10.1186/s13058-015-0660-6. Breast Cancer Res. 2016. PMID: 26728744 Free PMC article.
-
Expression of Bcl-2 family proteins and associated clinicopathologic factors predict survival outcome in patients with oral squamous cell carcinoma.Oral Oncol. 2009 Mar;45(3):225-33. doi: 10.1016/j.oraloncology.2008.05.021. Epub 2008 Aug 19. Oral Oncol. 2009. PMID: 18715811
Cited by
-
Recent advances in minimally invasive biomarkers of OSCC: from generalized to personalized approach.Front Oral Health. 2024 Aug 2;5:1426507. doi: 10.3389/froh.2024.1426507. eCollection 2024. Front Oral Health. 2024. PMID: 39157206 Free PMC article. Review.
-
Prediction of survival of HPV16-negative, p16-negative oral cavity cancer patients using a 13-gene signature: A multicenter study using FFPE samples.Oral Oncol. 2020 Jan;100:104487. doi: 10.1016/j.oraloncology.2019.104487. Epub 2019 Dec 10. Oral Oncol. 2020. PMID: 31835136 Free PMC article.
-
MTA3-SOX2 Module Regulates Cancer Stemness and Contributes to Clinical Outcomes of Tongue Carcinoma.Front Oncol. 2019 Aug 27;9:816. doi: 10.3389/fonc.2019.00816. eCollection 2019. Front Oncol. 2019. PMID: 31552166 Free PMC article.
-
A systematic review of proteomic biomarkers in oral squamous cell cancer.World J Surg Oncol. 2021 Oct 28;19(1):315. doi: 10.1186/s12957-021-02423-y. World J Surg Oncol. 2021. PMID: 34711249 Free PMC article.
-
Molecular Classification of Lymph Node Metastases Subtypes Predict for Survival in Head and Neck Cancer.Clin Cancer Res. 2019 Mar 15;25(6):1795-1808. doi: 10.1158/1078-0432.CCR-18-1884. Epub 2018 Dec 20. Clin Cancer Res. 2019. PMID: 30573692 Free PMC article.
References
-
- Japan Society for Head and Neck Cancer Registry Committee. Report of head and neck cancer registry of japan clinical statistic of registered patients, 2013. Jpn J Head Neck Cancer. 2015;41:1–115.
-
- Gibson MK, Forastiere AA. Multidisciplinary approaches in the management of advanced head and neck tumors: state of the art. Curr Opin Oncol. 2004;16:220–224. - PubMed
-
- Vermorken JB, Mesia R, Rivera F, Remenar E, Kawecki A, Rottey S, Erfan J, Zabolotnyy D, Kienzer HR, Cupissol D, Peyrade F, Benasso M, Vynnychenko I, et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med. 2008;359:1116–1127. doi: 10.1056/NEJMoa0802656. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

