Genetic analysis of porcine circovirus type 2 from pigs affected with PMWS in Chile reveals intergenotypic recombination
- PMID: 28978346
- PMCID: PMC5628495
- DOI: 10.1186/s12985-017-0850-1
Genetic analysis of porcine circovirus type 2 from pigs affected with PMWS in Chile reveals intergenotypic recombination
Abstract
Background: Porcine circovirus type 2 (PCV2) is a very small, non-enveloped and icosahedral virus, with circular single stranded DNA genome. This virus is the most ubiquitous and persistent pathogen currently affecting the swine industry worldwide. PCV2 has been implicated as the major causative agent of postweaning multisystemic wasting syndrome (PMWS), a disease which is characterized by severe immunosuppressive effects in the porcine host. Worldwide PCV2 isolates have been classified into four different genotypes, PCV2a, PCV2b, PCV2c and PCVd. The goal of this work was to conduct the first phylogenetic analysis of PCV2 in Chile.
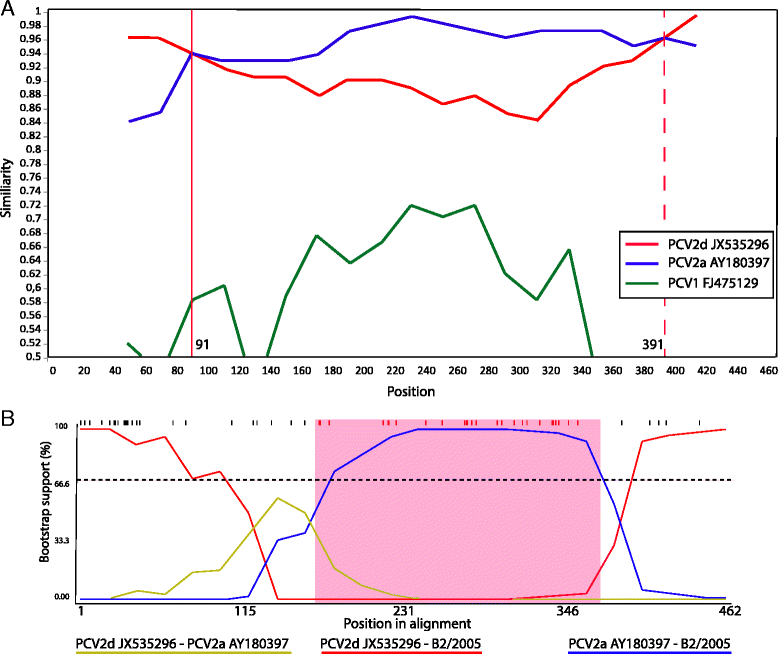
Methods: PCV2 partial ORF2 sequences (462 nt) obtained from 29 clinical cases of PMWS in 22 Chilean intensive swine farms, covering over the 90% of the local pork-production, were analyzed.
Results: 14% and 52% of sequences belonged to the genotypes PCV2a and PCV2b, respectively. Surprisingly, 34% of sequences were PCV2a/PCV2d recombinant viruses.
Conclusions: Our findings suggested that a novel cluster of Chilean sequences emerged resulting from intergenotypic recombination between PCV2a and PCV2d.
Keywords: Genetic diversity; PCV2; PMWS; Recombination.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


Similar articles
-
Porcine circovirus type 2 (PCV2): genetic variation and newly emerging genotypes in China.Virol J. 2010 Oct 19;7:273. doi: 10.1186/1743-422X-7-273. Virol J. 2010. PMID: 20955622 Free PMC article.
-
Genetic characterization of porcine circovirus type 2 in piglets from PMWS-affected and -negative farms in Thailand.Virol J. 2011 Feb 28;8:88. doi: 10.1186/1743-422X-8-88. Virol J. 2011. PMID: 21356069 Free PMC article.
-
A proposal on porcine circovirus type 2 (PCV2) genotype definition and their relation with postweaning multisystemic wasting syndrome (PMWS) occurrence.Vet Microbiol. 2008 Apr 1;128(1-2):23-35. doi: 10.1016/j.vetmic.2007.09.007. Epub 2007 Sep 19. Vet Microbiol. 2008. PMID: 17976930
-
Current state of knowledge on porcine circovirus type 2-associated lesions.Vet Pathol. 2013 Jan;50(1):23-38. doi: 10.1177/0300985812450726. Epub 2012 Jun 12. Vet Pathol. 2013. PMID: 22692624 Review.
-
Porcine circovirus type 2 and its associated diseases in Korea.Virus Res. 2012 Mar;164(1-2):107-13. doi: 10.1016/j.virusres.2011.10.013. Epub 2011 Oct 20. Virus Res. 2012. PMID: 22027190 Review.
Cited by
-
Novel influenza A viruses in pigs with zoonotic potential, Chile.Microbiol Spectr. 2024 Apr 2;12(4):e0218123. doi: 10.1128/spectrum.02181-23. Epub 2024 Mar 7. Microbiol Spectr. 2024. PMID: 38446039 Free PMC article.
-
Ubiquitous influenza A virus in Chilean swine before the H1N1pdm09 introduction.Transbound Emerg Dis. 2021 Nov;68(6):3174-3179. doi: 10.1111/tbed.14243. Epub 2021 Jul 30. Transbound Emerg Dis. 2021. PMID: 34288514 Free PMC article.
-
Genetic Diversity of Porcine Circovirus Isolated from Korean Wild Boars.Pathogens. 2020 Jun 9;9(6):457. doi: 10.3390/pathogens9060457. Pathogens. 2020. PMID: 32526932 Free PMC article.
-
Genomic composition and pathomechanisms of porcine circoviruses: A review.Virulence. 2024 Dec;15(1):2439524. doi: 10.1080/21505594.2024.2439524. Epub 2024 Dec 11. Virulence. 2024. PMID: 39662970 Free PMC article. Review.
-
A Heterologous Viral Protein Scaffold for Chimeric Antigen Design: An Example PCV2 Virus Vaccine Candidate.Viruses. 2020 Mar 31;12(4):385. doi: 10.3390/v12040385. Viruses. 2020. PMID: 32244384 Free PMC article.
References
-
- Allan G, Meehan B, Todd D, Kennedy S, McNeilly F, Ellis J, Clark EG, Harding J, Espuna E, Botner A, et al. Novel porcine circoviruses from pigs with wasting disease syndromes. Vet Rec. 1998;142(17):467–468. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

