MYC and MCL1 Cooperatively Promote Chemotherapy-Resistant Breast Cancer Stem Cells via Regulation of Mitochondrial Oxidative Phosphorylation
- PMID: 28978427
- PMCID: PMC5650077
- DOI: 10.1016/j.cmet.2017.09.009
MYC and MCL1 Cooperatively Promote Chemotherapy-Resistant Breast Cancer Stem Cells via Regulation of Mitochondrial Oxidative Phosphorylation
Abstract
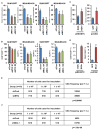
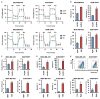
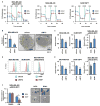
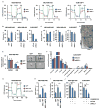
Most patients with advanced triple-negative breast cancer (TNBC) develop drug resistance. MYC and MCL1 are frequently co-amplified in drug-resistant TNBC after neoadjuvant chemotherapy. Herein, we demonstrate that MYC and MCL1 cooperate in the maintenance of chemotherapy-resistant cancer stem cells (CSCs) in TNBC. MYC and MCL1 increased mitochondrial oxidative phosphorylation (mtOXPHOS) and the generation of reactive oxygen species (ROS), processes involved in maintenance of CSCs. A mutant of MCL1 that cannot localize in mitochondria reduced mtOXPHOS, ROS levels, and drug-resistant CSCs without affecting the anti-apoptotic function of MCL1. Increased levels of ROS, a by-product of activated mtOXPHOS, led to the accumulation of HIF-1α. Pharmacological inhibition of HIF-1α attenuated CSC enrichment and tumor initiation in vivo. These data suggest that (1) MYC and MCL1 confer resistance to chemotherapy by expanding CSCs via mtOXPHOS and (2) targeting mitochondrial respiration and HIF-1α may reverse chemotherapy resistance in TNBC.
Keywords: MCL1; MYC; cancer stem cell; chemotherapy resistance; mitochondrial respiration; triple negative breast cancer.
Copyright © 2017. Published by Elsevier Inc.
Figures


References
-
- Alirol E, Martinou JC. Mitochondria and cancer: is there a morphological connection? Oncogene. 2006;25:4706–4716. - PubMed
-
- Balko JM, Giltnane JM, Wang K, Schwarz LJ, Young CD, Cook RS, Owens P, Sanders ME, Kuba MG, Sanchez V, et al. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov. 2014;4:232–245. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

