Foxp1 in Forebrain Pyramidal Neurons Controls Gene Expression Required for Spatial Learning and Synaptic Plasticity
- PMID: 28978667
- PMCID: PMC5678023
- DOI: 10.1523/JNEUROSCI.1005-17.2017
Foxp1 in Forebrain Pyramidal Neurons Controls Gene Expression Required for Spatial Learning and Synaptic Plasticity
Erratum in
-
Correction: Araujo et al., "Foxp1 in Forebrain Pyramidal Neurons Controls Gene Expression Required for Spatial Learning and Synaptic Plasticity".J Neurosci. 2018 Mar 21;38(12):3147. doi: 10.1523/JNEUROSCI.0293-18.2018. J Neurosci. 2018. PMID: 31329677 Free PMC article.
Abstract
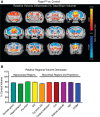
Genetic perturbations of the transcription factor Forkhead Box P1 (FOXP1) are causative for severe forms of autism spectrum disorder that are often comorbid with intellectual disability. Recent work has begun to reveal an important role for FoxP1 in brain development, but the brain-region-specific contributions of Foxp1 to autism and intellectual disability phenotypes have yet to be determined fully. Here, we describe Foxp1 conditional knock-out (Foxp1cKO) male and female mice with loss of Foxp1 in the pyramidal neurons of the neocortex and the CA1/CA2 subfields of the hippocampus. Foxp1cKO mice exhibit behavioral phenotypes that are of potential relevance to autism spectrum disorder, including hyperactivity, increased anxiety, communication impairments, and decreased sociability. In addition, Foxp1cKO mice have gross deficits in learning and memory tasks of relevance to intellectual disability. Using a genome-wide approach, we identified differentially expressed genes in the hippocampus of Foxp1cKO mice associated with synaptic function and development. Furthermore, using magnetic resonance imaging, we uncovered a significant reduction in the volumes of both the entire hippocampus as well as individual hippocampal subfields of Foxp1cKO mice. Finally, we observed reduced maintenance of LTP in area CA1 of the hippocampus in these mutant mice. Together, these data suggest that proper expression of Foxp1 in the pyramidal neurons of the forebrain is important for regulating gene expression pathways that contribute to specific behaviors reminiscent of those seen in autism and intellectual disability. In particular, Foxp1 regulation of gene expression appears to be crucial for normal hippocampal development, CA1 plasticity, and spatial learning.SIGNIFICANCE STATEMENT Loss-of-function mutations in the transcription factor Forkhead Box P1 (FOXP1) lead to autism spectrum disorder and intellectual disability. Understanding the potential brain-region-specific contributions of FOXP1 to disease-relevant phenotypes could be a critical first step in the management of patients with these mutations. Here, we report that Foxp1 conditional knock-out (Foxp1cKO) mice with loss of Foxp1 in the neocortex and hippocampus display autism and intellectual-disability-relevant behaviors. We also show that these phenotypes correlate with changes in both the genomic and physiological profiles of the hippocampus in Foxp1cKO mice. Our work demonstrates that brain-region-specific FOXP1 expression may relate to distinct, clinically relevant phenotypes.
Keywords: autism; gene expression; hippocampus; spatial learning; synaptic plasticity.
Copyright © 2017 the authors 0270-6474/17/3710918-15$15.00/0.
Figures
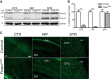
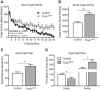
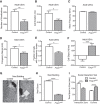


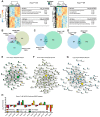

References
-
- Andrews S. (2010) FastQC: a quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
-
- Araujo DJ, Anderson AG, Berto S, Runnels W, Harper M, Ammanuel S, Rieger MA, Huang HC, Rajkovich K, Loerwald KW, Dekker JD, Tucker HO, Dougherty JD, Gibson JR, Konopka G (2015) FoxP1 orchestration of ASD-relevant signaling pathways in the striatum. Genes Dev 29:2081–2096. 10.1101/gad.267989.115 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
