Human Metapneumovirus Induces Formation of Inclusion Bodies for Efficient Genome Replication and Transcription
- PMID: 28978704
- PMCID: PMC5709606
- DOI: 10.1128/JVI.01282-17
Human Metapneumovirus Induces Formation of Inclusion Bodies for Efficient Genome Replication and Transcription
Abstract
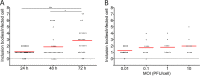
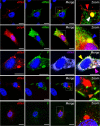
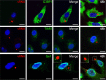
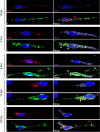
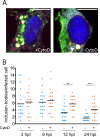
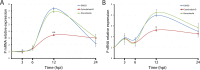
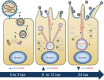
Human metapneumovirus (HMPV) causes significant upper and lower respiratory disease in all age groups worldwide. The virus possesses a negative-sense single-stranded RNA genome of approximately 13.3 kb encapsidated by multiple copies of the nucleoprotein (N), giving rise to helical nucleocapsids. In addition, copies of the phosphoprotein (P) and the large RNA polymerase (L) decorate the viral nucleocapsids. After viral attachment, endocytosis, and fusion mediated by the viral glycoproteins, HMPV nucleocapsids are released into the cell cytoplasm. To visualize the subsequent steps of genome transcription and replication, a fluorescence in situ hybridization (FISH) protocol was established to detect different viral RNA subpopulations in infected cells. The FISH probes were specific for detection of HMPV positive-sense RNA (+RNA) and viral genomic RNA (vRNA). Time course analysis of human bronchial epithelial BEAS-2B cells infected with HMPV revealed the formation of inclusion bodies (IBs) from early times postinfection. HMPV IBs were shown to be cytoplasmic sites of active transcription and replication, with the translation of viral proteins being closely associated. Inclusion body formation was consistent with an actin-dependent coalescence of multiple early replicative sites. Time course quantitative reverse transcription-PCR analysis suggested that the coalescence of inclusion bodies is a strategy to efficiently replicate and transcribe the viral genome. These results provide a better understanding of the steps following HMPV entry and have important clinical implications.IMPORTANCE Human metapneumovirus (HMPV) is a recently discovered pathogen that affects human populations of all ages worldwide. Reinfections are common throughout life, but no vaccines or antiviral treatments are currently available. In this work, a spatiotemporal analysis of HMPV replication and transcription in bronchial epithelial cell-derived immortal cells was performed. HMPV was shown to induce the formation of large cytoplasmic granules, named inclusion bodies, for genome replication and transcription. Unlike other cytoplasmic structures, such as stress granules and processing bodies, inclusion bodies are exclusively present in infected cells and contain HMPV RNA and proteins to more efficiently transcribe and replicate the viral genome. Though inclusion body formation is nuanced, it corresponds to a more generalized strategy used by different viruses, including filoviruses and rhabdoviruses, for genome transcription and replication. Thus, an understanding of inclusion body formation is crucial for the discovery of innovative therapeutic targets.
Keywords: HMPV; inclusion bodies; pneumovirus; replication.
Copyright © 2017 American Society for Microbiology.
Figures



References
-
- Boivin G, Abed Y, Pelletier G, Ruel L, Moisan D, Cote S, Peret TC, Erdman DD, Anderson LJ. 2002. Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory-tract infections in all age groups. J Infect Dis 186:1330–1334. doi: 10.1086/344319. - DOI - PubMed
-
- Afonso CL, Amarasinghe GK, Banyai K, Bao Y, Basler CF, Bavari S, Bejerman N, Blasdell KR, Briand FX, Briese T, Bukreyev A, Calisher CH, Chandran K, Cheng J, Clawson AN, Collins PL, Dietzgen RG, Dolnik O, Domier LL, Durrwald R, Dye JM, Easton AJ, Ebihara H, Farkas SL, Freitas-Astua J, Formenty P, Fouchier RA, Fu Y, Ghedin E, Goodin MM, Hewson R, Horie M, Hyndman TH, Jiang D, Kitajima EW, Kobinger GP, Kondo H, Kurath G, Lamb RA, Lenardon S, Leroy EM, Li CX, Lin XD, Liu L, Longdon B, Marton S, Maisner A, Muhlberger E, Netesov SV, Nowotny N, et al. 2016. Taxonomy of the order Mononegavirales: update 2016. Arch Virol 161:2351–2360. doi: 10.1007/s00705-016-2880-1. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

