Functional Stability of HIV-1 Envelope Trimer Affects Accessibility to Broadly Neutralizing Antibodies at Its Apex
- PMID: 28978711
- PMCID: PMC5709597
- DOI: 10.1128/JVI.01216-17
Functional Stability of HIV-1 Envelope Trimer Affects Accessibility to Broadly Neutralizing Antibodies at Its Apex
Abstract
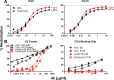
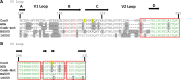
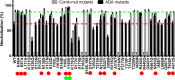
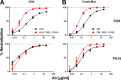
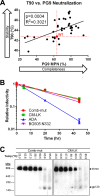
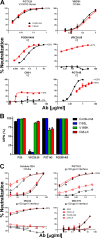
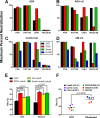
The trimeric envelope glycoprotein spike (Env) of HIV-1 is the target of vaccine development to elicit broadly neutralizing antibodies (bnAbs). Env trimer instability and heterogeneity in principle make subunit interfaces inconsistent targets for the immune response. Here, we investigate how functional stability of Env relates to neutralization sensitivity to V2 bnAbs and V3 crown antibodies that engage subunit interfaces upon binding to unliganded Env. Env heterogeneity was inferred when antibodies neutralized a mutant Env with a plateau of less than 100% neutralization. A statistically significant correlation was found between the stability of mutant Envs and the MPN of V2 bnAb, PG9, as well as an inverse correlation between stability of Env and neutralization by V3 crown antibody, 447-52D. A number of Env-stabilizing mutations and V2 bnAb-enhancing mutations were identified in Env, but they did not always overlap, indicating distinct requirements of functional stabilization versus antibody recognition. Blocking complex glycosylation of Env affected V2 bnAb recognition, as previously described, but also notably increased functional stability of Env. This study shows how instability and heterogeneity affect antibody sensitivity of HIV-1 Env, which is relevant to vaccine design involving its dynamic apex.IMPORTANCE The Env trimer is the only viral protein on the surface of HIV-1 and is the target of neutralizing antibodies that reduce viral infectivity. Quaternary epitopes at the apex of the spike are recognized by some of the most potent and broadly neutralizing antibodies to date. Being that their glycan-protein hybrid epitopes are at subunit interfaces, the resulting heterogeneity can lead to partial neutralization. Here, we screened for mutations in Env that allowed for complete neutralization by the bnAbs. We found that when mutations outside V2 increased V2 bnAb recognition, they often also increased Env stability-of-function and decreased binding by narrowly neutralizing antibodies to the V3 crown. Three mutations together increased neutralization by V2 bnAb and eliminated binding by V3 crown antibodies. These results may aid the design of immunogens that elicit antibodies to the trimer apex.
Keywords: envelope; gp120; gp41; human immunodeficiency virus; neutralizing antibodies; protein stability; vaccines.
Copyright © 2017 American Society for Microbiology.
Figures



References
-
- Burton DR, Ahmed R, Barouch DH, Butera ST, Crotty S, Godzik A, Kaufmann DE, McElrath MJ, Nussenzweig MC, Pulendran B, Scanlan CN, Schief WR, Silvestri G, Streeck H, Walker BD, Walker LM, Ward AB, Wilson IA, Wyatt R. 2012. A blueprint for HIV vaccine discovery. Cell Host Microbe 12:396–407. doi:10.1016/j.chom.2012.09.008. - DOI - PMC - PubMed
-
- Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, Arendt H, Kim HJ, Lee JH, Pugach P, Williams M, Debnath G, Moldt B, van Breemen MJ, Isik G, Medina-Ramirez M, Back JW, Koff WC, Julien JP, Rakasz EG, Seaman MS, Guttman M, Lee KK, Klasse PJ, LaBranche C, Schief WR, Wilson IA, Overbaugh J, Burton DR, Ward AB, Montefiori DC, Dean H, Moore JP. 2015. HIV-1 VACCINES: HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 349:aac4223. doi:10.1126/science.aac4223. - DOI - PMC - PubMed
-
- McCoy LE, Falkowska E, Doores KJ, Le K, Sok D, van Gils MJ, Euler Z, Burger JA, Seaman MS, Sanders RW, Schuitemaker H, Poignard P, Wrin T, Burton DR. 2015. Incomplete neutralization and deviation from sigmoidal neutralization curves for HIV broadly neutralizing monoclonal antibodies. PLoS Pathog 11:e1005110. doi:10.1371/journal.ppat.1005110. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

