T Cell-Macrophage Fusion Triggers Multinucleated Giant Cell Formation for HIV-1 Spreading
- PMID: 28978713
- PMCID: PMC5709600
- DOI: 10.1128/JVI.01237-17
T Cell-Macrophage Fusion Triggers Multinucleated Giant Cell Formation for HIV-1 Spreading
Abstract
HIV-1-infected macrophages participate in virus dissemination and establishment of virus reservoirs in host tissues, but the mechanisms for virus cell-to-cell transfer to macrophages remain unknown. Here, we reveal the mechanisms for cell-to-cell transfer from infected T cells to macrophages and virus spreading between macrophages. We show that contacts between infected T lymphocytes and macrophages lead to cell fusion for the fast and massive transfer of CCR5-tropic viruses to macrophages. Through the merge of viral material between T cells and macrophages, these newly formed lymphocyte-macrophage fused cells acquire the ability to fuse with neighboring noninfected macrophages. Together, these two-step envelope-dependent cell fusion processes lead to the formation of highly virus-productive multinucleated giant cells reminiscent of the infected multinucleated giant macrophages detected in HIV-1-infected patients and simian immunodeficiency virus-infected macaques. These mechanisms represent an original mode of virus transmission for viral spreading and a new model for the formation of macrophage virus reservoirs during infection.IMPORTANCE We reveal a very efficient mechanism involved in cell-to-cell transfer from infected T cells to macrophages and subsequent virus spreading between macrophages by a two-step cell fusion process. Infected T cells first establish contacts and fuse with macrophage targets. The newly formed lymphocyte-macrophage fused cells then acquire the ability to fuse with surrounding uninfected macrophages, leading to the formation of infected multinucleated giant cells that can survive for a long time, as evidenced in vivo in lymphoid organs and the central nervous system. This route of infection may be a major determinant for virus dissemination and the formation of macrophage virus reservoirs in host tissues during HIV-1 infection.
Keywords: HIV-1; T lymphocytes; cell fusion; cell-to-cell transfer; macrophages.
Copyright © 2017 American Society for Microbiology.
Figures
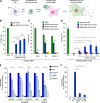



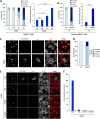
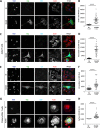

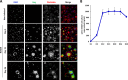

Similar articles
-
Cell-to-Cell Spreading of HIV-1 in Myeloid Target Cells Escapes SAMHD1 Restriction.mBio. 2019 Nov 19;10(6):e02457-19. doi: 10.1128/mBio.02457-19. mBio. 2019. PMID: 31744918 Free PMC article.
-
Loss of a conserved N-linked glycosylation site in the simian immunodeficiency virus envelope glycoprotein V2 region enhances macrophage tropism by increasing CD4-independent cell-to-cell transmission.J Virol. 2014 May;88(9):5014-28. doi: 10.1128/JVI.02785-13. Epub 2014 Feb 19. J Virol. 2014. PMID: 24554659 Free PMC article.
-
Quantitation of Productively Infected Monocytes and Macrophages of Simian Immunodeficiency Virus-Infected Macaques.J Virol. 2016 May 27;90(12):5643-5656. doi: 10.1128/JVI.00290-16. Print 2016 Jun 15. J Virol. 2016. PMID: 27030272 Free PMC article.
-
The macrophage in HIV infection.Immunobiology. 2001 Dec;204(5):598-602. doi: 10.1078/0171-2985-00098. Immunobiology. 2001. PMID: 11846224 Review.
-
HIV Persistence in Adipose Tissue Reservoirs.Curr HIV/AIDS Rep. 2018 Feb;15(1):60-71. doi: 10.1007/s11904-018-0378-z. Curr HIV/AIDS Rep. 2018. PMID: 29423731 Free PMC article. Review.
Cited by
-
Cellular HIV Reservoirs and Viral Rebound from the Lymphoid Compartments of 4'-Ethynyl-2-Fluoro-2'-Deoxyadenosine (EFdA)-Suppressed Humanized Mice.Viruses. 2019 Mar 13;11(3):256. doi: 10.3390/v11030256. Viruses. 2019. PMID: 30871222 Free PMC article.
-
Mechanisms of HIV-1 cell-to-cell transmission and the establishment of the latent reservoir.Virus Res. 2019 May;265:115-121. doi: 10.1016/j.virusres.2019.03.014. Epub 2019 Mar 21. Virus Res. 2019. PMID: 30905686 Free PMC article. Review.
-
Kinetic Analysis of the Motility of Giant Virus-Infected Amoebae Using Phase-Contrast Microscopic Images.Front Microbiol. 2020 Jan 17;10:3014. doi: 10.3389/fmicb.2019.03014. eCollection 2019. Front Microbiol. 2020. PMID: 32038516 Free PMC article.
-
Cellular and molecular actors of myeloid cell fusion: podosomes and tunneling nanotubes call the tune.Cell Mol Life Sci. 2021 Sep;78(17-18):6087-6104. doi: 10.1007/s00018-021-03875-x. Epub 2021 Jul 23. Cell Mol Life Sci. 2021. PMID: 34296319 Free PMC article. Review.
-
Vpr Is a VIP: HIV Vpr and Infected Macrophages Promote Viral Pathogenesis.Viruses. 2020 Jul 27;12(8):809. doi: 10.3390/v12080809. Viruses. 2020. PMID: 32726944 Free PMC article. Review.
References
-
- Harbison C, Zhuang K, Gettie A, Blanchard J, Knight H, Didier P, Cheng-Mayer C, Westmoreland S. 2014. Giant cell encephalitis and microglial infection with mucosally transmitted simian-human immunodeficiency virus SHIVSF162P3N in rhesus macaques. J Neurovirol 20:62–72. doi:10.1007/s13365-013-0229-z. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

