Unraveling Macrophage Heterogeneity in Erythroblastic Islands
- PMID: 28979259
- PMCID: PMC5611421
- DOI: 10.3389/fimmu.2017.01140
Unraveling Macrophage Heterogeneity in Erythroblastic Islands
Abstract
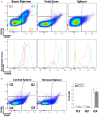
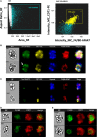
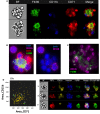
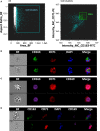
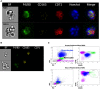
Mammalian erythropoiesis occurs within erythroblastic islands (EBIs), niches where maturing erythroblasts interact closely with a central macrophage. While it is generally accepted that EBI macrophages play an important role in erythropoiesis, thorough investigation of the mechanisms by which they support erythropoiesis is limited largely by inability to identify and isolate the specific macrophage sub-population that constitute the EBI. Early studies utilized immunohistochemistry or immunofluorescence to study EBI morphology and structure, while more recent efforts have used flow cytometry for high-throughput quantitative characterization of EBIs and their central macrophages. However, these approaches based on the expectation that EBI macrophages are a homogeneous population (F4/80+/CD169+/VCAM-1+ for example) provide an incomplete picture and potentially overlook critical information about the nature and biology of the islands and their central macrophages. Here, we present a novel method for analysis of EBI macrophages from hematopoietic tissues of mice and rats using multispectral imaging flow cytometry (IFC), which combines the high-throughput advantage of flow cytometry with the morphological and fluorescence features derived from microscopy. This method provides both quantitative analysis of EBIs, as well as structural and morphological details of the central macrophages and associated cells. Importantly, the images, combined with quantitative software features, can be used to evaluate co-expression of phenotypic markers which is crucial since some antigens used to identify macrophages (e.g., F4/80 and CD11b) can be expressed on non-erythroid cells associated with the islands instead of, or in addition to the central macrophage itself. We have used this method to analyze native EBIs from different hematopoietic tissues and evaluated the expression of several markers that have been previously reported to be expressed on EBI macrophages. We found that VCAM-1, F4/80, and CD169 are expressed heterogeneously by the central macrophages within the EBIs, while CD11b, although abundantly expressed by cells within the islands, is not expressed on the EBI macrophages. Moreover, differences in the phenotype of EBIs in rats compared to mice point to potential functional differences between these species. These data demonstrate the usefulness of IFC in analysis and characterization of EBIs and more importantly in exploring the heterogeneity and plasticity of EBI macrophages.
Keywords: CD11b; CD163; CD169; VCAM-1; erythroblastic islands; erythropoiesis; imaging flow cytometry; macrophages.
Figures


References
-
- Van Putten LM. The life span of red cells in the rat and the mouse as determined by labeling with DFP32 in vivo. Blood (1958) 13(8):789–94. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

