Quantitative proteomics analysis of mitochondrial proteins in lung adenocarcinomas and normal lung tissue using iTRAQ and tandem mass spectrometry
- PMID: 28979670
- PMCID: PMC5622239
Quantitative proteomics analysis of mitochondrial proteins in lung adenocarcinomas and normal lung tissue using iTRAQ and tandem mass spectrometry
Abstract
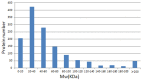
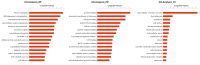
Lung adenocarcinoma is the most common type of lung cancer. Unfortunately, lung adenocarcinoma has a poor prognosis and the pathogenesis remains unclear. Mitochondria are important mediators of tumorigenesis. However, the proteomics profile of lung adenocarcinoma mitochondrial proteins has not been elucidated. In this study, we investigated differences in the mitochondrial protein profiles between lung adenocarcinomas and normal tissue. Laser capture microdissection (LCM) was used to isolate the target cells from lung adenocarcinomas and normal tissue. The differential expression of mitochondrial proteins was determined using isobaric tags for relative and absolute quantitation (iTRAQ) combined with two-dimensional liquid chromatography-tandem mass spectrometry (2D-LC-MS/MS). Bioinformatics analysis was performed using Gene Ontology and KEGG databases. As a result, 510 differentially expressed proteins were identified, 315 of which were upregulated and 195 that were downregulated. Of these proteins, 35.5% were mitochondrial or mitochondrial-related and were involved in binding, catalysis, molecular transduction, transport, and molecular structure. Based on the differentially expressed proteins, 63 pathways were significantly enriched through KEGG. The overexpression and cellular distribution of the mitochondrial protein C1QBP in the lung cancer samples was confirmed and verified by Western blotting. The relationship between C1QBP expression and clinicopathological features in lung cancer patients was likewise evaluated using immunohistochemistry, which revealed that the upregulation of C1QBP was associated with lymph node metastasis, pathological grade and clinical stage of TNM. The results indicate that the iTRAQ 2D-LC-MS/MS technique is a potential method for comparing mitochondrial protein profiles between tumor and normal tissue and could aid in identifying novel biomarkers and the mechanisms underlying carcinogenesis.
Keywords: Lung adenocarcinoma; iTRAQ; mitochondrial protein; proteomic profile.
Conflict of interest statement
None.
Figures







Similar articles
-
Comparative membrane proteomic analysis between lung adenocarcinoma and normal tissue by iTRAQ labeling mass spectrometry.Am J Transl Res. 2014 May 15;6(3):267-80. eCollection 2014. Am J Transl Res. 2014. PMID: 24936219 Free PMC article.
-
Identification and Verification of Two Novel Differentially Expressed Proteins from Non-neoplastic Mucosa and Colorectal Carcinoma Via iTRAQ Combined with Liquid Chromatography-Mass Spectrometry.Pathol Oncol Res. 2020 Apr;26(2):967-976. doi: 10.1007/s12253-019-00651-y. Epub 2019 Mar 29. Pathol Oncol Res. 2020. PMID: 30927204
-
ITRAQ-Based Proteomics Analysis of Triptolide On Human A549 Lung Adenocarcinoma Cells.Cell Physiol Biochem. 2018;45(3):917-934. doi: 10.1159/000487286. Epub 2018 Feb 2. Cell Physiol Biochem. 2018. PMID: 29428961
-
Exploration of differentially expressed plasma proteins in patients with lung adenocarcinoma using iTRAQ-coupled 2D LC-MS/MS.Clin Respir J. 2018 Jun;12(6):2036-2045. doi: 10.1111/crj.12771. Epub 2018 Feb 21. Clin Respir J. 2018. PMID: 29363881
-
Identification of integrin β1 as a prognostic biomarker for human lung adenocarcinoma using 2D-LC-MS/MS combined with iTRAQ technology.Oncol Rep. 2013 Jul;30(1):341-9. doi: 10.3892/or.2013.2477. Epub 2013 May 15. Oncol Rep. 2013. PMID: 23677397
Cited by
-
Comparative mitochondrial proteomic analysis of human large cell lung cancer cell lines with different metastasis potential.Thorac Cancer. 2019 May;10(5):1111-1128. doi: 10.1111/1759-7714.13052. Epub 2019 Apr 4. Thorac Cancer. 2019. PMID: 30950202 Free PMC article.
-
Differentiation-related genes in tumor-associated macrophages as potential prognostic biomarkers in non-small cell lung cancer.Front Immunol. 2023 Mar 9;14:1123840. doi: 10.3389/fimmu.2023.1123840. eCollection 2023. Front Immunol. 2023. PMID: 36969247 Free PMC article.
-
Exploring potential biomarkers for lung adenocarcinoma using LC-MS/MS metabolomics.J Int Med Res. 2020 Apr;48(4):300060519897215. doi: 10.1177/0300060519897215. J Int Med Res. 2020. PMID: 32316791 Free PMC article.
-
Hypoxia-Mediated Complement 1q Binding Protein Regulates Metastasis and Chemoresistance in Triple-Negative Breast Cancer and Modulates the PKC-NF-κB-VCAM-1 Signaling Pathway.Front Cell Dev Biol. 2021 Feb 23;9:607142. doi: 10.3389/fcell.2021.607142. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33708767 Free PMC article.
-
Potential biomarkers and targets of mitochondrial dynamics.Clin Transl Med. 2021 Aug;11(8):e529. doi: 10.1002/ctm2.529. Clin Transl Med. 2021. PMID: 34459143 Free PMC article. Review.
References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. - PubMed
-
- Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. - PubMed
-
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
