Identification of Chlamydomonas reinhardtii endogenous genic flanking sequences for improved transgene expression
- PMID: 28980350
- PMCID: PMC5718938
- DOI: 10.1111/tpj.13731
Identification of Chlamydomonas reinhardtii endogenous genic flanking sequences for improved transgene expression
Abstract
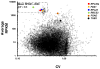
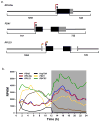
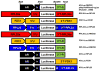
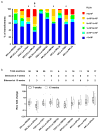
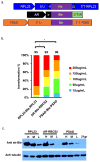
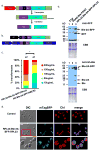
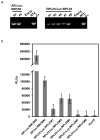
Chlamydomonas reinhardtii is a unicellular green alga that has attracted interest due to its potential biotechnological applications, and as a model for algal biofuel and energy metabolism. Despite all the advantages that this unicellular alga offers, poor and inconsistent expression of nuclear transgenes remains an obstacle for basic and applied research. We used a data-mining strategy to identify highly expressed genes in Chlamydomonas whose flanking sequences were tested for the ability to drive heterologous nuclear transgene expression. Candidates identified in this search included two ribosomal protein genes, RPL35a and RPL23, and ferredoxin, FDX1, whose flanking regions including promoters, terminators and untranslated sequences could drive stable luciferase transgene expression to significantly higher levels than the commonly used Hsp70A-RBCS2 (AR) hybrid promoter/terminator sequences. The RPL23 flanking sequences were further tested using the zeocin resistance gene sh-ble as a reporter in monocistronic and dicistronic constructs, and consistently yielded higher numbers of zeocin-resistant transformants and higher levels of resistance than AR- or PSAD-based vectors. Chlamydomonas RPL23 sequences also enabled transgene expression in Volvox carteri. Our study provides an additional benchmark for strong constitutive expression of transgenes in Chlamydomonas, and develops a general approach for identifying flanking sequences that can be used to drive transgene expression for any organism where transcriptome data are available.
Keywords: Chlamydomonas reinhardtii; Volvox carteri; dicistronic vector; luciferase; promoter; skipping peptide; technical advance; terminator; untranslated region.
© 2017 The Authors The Plant Journal © 2017 John Wiley & Sons Ltd.
Figures
Similar articles
-
Swapped green algal promoters: aphVIII-based gene constructs with Chlamydomonas flanking sequences work as dominant selectable markers in Volvox and vice versa.Plant Cell Rep. 2006 Jun;25(6):582-91. doi: 10.1007/s00299-006-0121-x. Epub 2006 Feb 3. Plant Cell Rep. 2006. PMID: 16456645
-
Stable nuclear transformation of Gonium pectorale.BMC Biotechnol. 2009 Jul 10;9:64. doi: 10.1186/1472-6750-9-64. BMC Biotechnol. 2009. PMID: 19591675 Free PMC article.
-
Stable nuclear transformation of Eudorina elegans.BMC Biotechnol. 2013 Feb 12;13:11. doi: 10.1186/1472-6750-13-11. BMC Biotechnol. 2013. PMID: 23402598 Free PMC article.
-
Good News for Nuclear Transgene Expression in Chlamydomonas.Cells. 2019 Nov 28;8(12):1534. doi: 10.3390/cells8121534. Cells. 2019. PMID: 31795196 Free PMC article. Review.
-
[The regulation of erythropoietin gene expression].Sheng Li Ke Xue Jin Zhan. 1998 Jan;29(1):87-9. Sheng Li Ke Xue Jin Zhan. 1998. PMID: 12501716 Review. Chinese. No abstract available.
Cited by
-
Introduction of a leaky stop codon as molecular tool in Chlamydomonas reinhardtii.PLoS One. 2020 Aug 20;15(8):e0237405. doi: 10.1371/journal.pone.0237405. eCollection 2020. PLoS One. 2020. PMID: 32817702 Free PMC article.
-
Current Nuclear Engineering Strategies in the Green Microalga Chlamydomonas reinhardtii.Life (Basel). 2023 Jul 15;13(7):1566. doi: 10.3390/life13071566. Life (Basel). 2023. PMID: 37511941 Free PMC article. Review.
-
Molecular Advancements Establishing Chlamydomonas as a Host for Biotechnological Exploitation.Front Plant Sci. 2022 Jun 29;13:911483. doi: 10.3389/fpls.2022.911483. eCollection 2022. Front Plant Sci. 2022. PMID: 35845675 Free PMC article. Review.
-
Site-Specific Gene Knock-Out and On-Site Heterologous Gene Overexpression in Chlamydomonas reinhardtii via a CRISPR-Cas9-Mediated Knock-in Method.Front Plant Sci. 2020 Mar 20;11:306. doi: 10.3389/fpls.2020.00306. eCollection 2020. Front Plant Sci. 2020. PMID: 32265959 Free PMC article.
-
Advanced pathway engineering for phototrophic putrescine production.Plant Biotechnol J. 2022 Oct;20(10):1968-1982. doi: 10.1111/pbi.13879. Epub 2022 Jul 22. Plant Biotechnol J. 2022. PMID: 35748533 Free PMC article.
References
-
- Berthold P, Schmitt R, Mages W. An engineered Streptomyces hygroscopicus aph 7″ gene mediates dominant resistance against hygromycin B in Chlamydomonas reinhardtii. Protist. 2002;153:401–412. - PubMed
-
- Boynton JE, Gillham NW, Harris EH, et al. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science. 1988;240:1534–1538. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

