Neutral evolution of drug resistant colorectal cancer cell populations is independent of their KRAS status
- PMID: 28981524
- PMCID: PMC5628783
- DOI: 10.1371/journal.pone.0175484
Neutral evolution of drug resistant colorectal cancer cell populations is independent of their KRAS status
Abstract
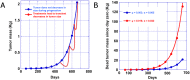
Emergence of tumor resistance to an anti-cancer therapy directed against a putative target raises several questions including: (1) do mutations in the target/pathway confer resistance? (2) Are these mutations pre-existing? (3) What is the relative fitness of cells with/without the mutation? We addressed these questions in patients with metastatic colorectal cancer (mCRC). We conducted an exhaustive review of published data to establish a median doubling time for CRCs and stained a cohort of CRCs to document mitotic indices. We analyzed published data and our own data to calculate rates of growth (g) and regression (d, decay) of tumors in patients with CRC correlating these results with the detection of circulating MT-KRAS DNA. Additionally we estimated mathematically the caloric burden of such tumors using data on mitotic and apoptotic indices. We conclude outgrowth of cells harboring intrinsic or acquired MT-KRAS cannot explain resistance to anti-EGFR (epidermal growth factor receptor) antibodies. Rates of tumor growth with panitumumab are unaffected by presence/absence of MT-KRAS. While MT-KRAS cells may be resistant to anti-EGFR antibodies, WT-KRAS cells also rapidly bypass this blockade suggesting inherent resistance mechanisms are responsible and a neutral evolution model is most appropriate. Using the above clinical data on tumor doubling times and mitotic and apoptotic indices we estimated the caloric intake required to support tumor growth and suggest it may explain in part cancer-associated cachexia.
Conflict of interest statement
Figures



References
-
- Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001; 293:876–80. doi: 10.1126/science.1062538 - DOI - PubMed
-
- Buczek M, Escudier B, Bartnik E, Szczylik C, Czarnecka A. Resistance to tyrosine kinase inhibitors in clear cell renal cell carcinoma: from the patient's bed to molecular mechanisms. Biochim Biophys Acta. 2014; 1845:31–41. doi: 10.1016/j.bbcan.2013.10.001 - DOI - PubMed
-
- von Bubnoff N, Schneller F, Peschel C, Duyster J. BCR-ABL gene mutations in relation to clinical resistance of Philadelphia-chromosome-positive leukaemia to STI571: a prospective study. Lancet. 2002; 359:487–91. doi: 10.1016/S0140-6736(02)07679-1 - DOI - PubMed
-
- Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005; 2:e73 doi: 10.1371/journal.pmed.0020073 - DOI - PMC - PubMed
-
- Hegedus C, Ozvegy-Laczka C, Apati A, Magócsi M, Német K, Orfi L, et al. Interaction of nilotinib, dasatinib and bosutinib with ABCB1 and ABCG2: implications for altered anti-cancer effects and pharmacological properties. Br J Pharmacol. 2009; 158:1153–64, 2009. doi: 10.1111/j.1476-5381.2009.00383.x - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

