NIMA-related kinase 1 (NEK1) regulates meiosis I spindle assembly by altering the balance between α-Adducin and Myosin X
- PMID: 28982183
- PMCID: PMC5628868
- DOI: 10.1371/journal.pone.0185780
NIMA-related kinase 1 (NEK1) regulates meiosis I spindle assembly by altering the balance between α-Adducin and Myosin X
Abstract
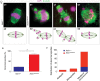
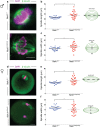
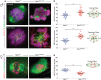
NIMA-related kinase 1 (NEK1) is a serine/threonine and tyrosine kinase that is highly expressed in mammalian germ cells. Mutations in Nek1 induce anemia, polycystic kidney and infertility. In this study we evaluated the role of NEK1 in meiotic spindle formation in both male and female gametes. Our results show that the lack of NEK1 provokes an abnormal organization of the meiosis I spindle characterized by elongated and/or multipolar spindles, and abnormal chromosome congression. The aberrant spindle structure is concomitant with the disruption in localization and protein levels of myosin X (MYO10) and α-adducin (ADD1), both of which are implicated in the regulation of spindle formation during mitosis. Interaction of ADD1 with MYO10 is dependent on phosphorylation, whereby phosphorylation of ADD1 enables its binding to MYO10 on mitotic spindles. Reduction in ADD1 protein in NEK1 mutant mice is associated with hyperphosphorylation of ADD1, thereby preventing the interaction with MYO10 during meiotic spindle formation. Our results reveal a novel regulatory role for NEK1 in the regulation of spindle architecture and function during meiosis.
Conflict of interest statement
Figures



References
-
- Gray S, Cohen PE. Control of Meiotic Crossovers: From Double-Strand Break Formation to Designation. Annu Rev Genet 2016;50: 175–210. doi: 10.1146/annurev-genet-120215-035111 - DOI - PMC - PubMed
-
- Duro E, Marston AL. From equator to pole: splitting chromosomes in mitosis and meiosis. Genes Dev 2015;29: 109–122. doi: 10.1101/gad.255554.114 - DOI - PMC - PubMed
-
- Bennabi I, Terret ME, Verlhac MH. Meiotic spindle assembly and chromosome segregation in oocytes. J Cell Biol 2016;215: 611–619. doi: 10.1083/jcb.201607062 - DOI - PMC - PubMed
-
- McKim KS, Hawley RS. Chromosomal control of meiotic cell division. Science 1995;270: 1595–1601. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

