Non-AUG translation: a new start for protein synthesis in eukaryotes
- PMID: 28982758
- PMCID: PMC5666671
- DOI: 10.1101/gad.305250.117
Non-AUG translation: a new start for protein synthesis in eukaryotes
Abstract
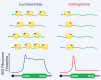
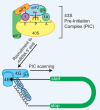
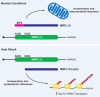
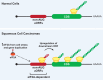
Although it was long thought that eukaryotic translation almost always initiates at an AUG start codon, recent advancements in ribosome footprint mapping have revealed that non-AUG start codons are used at an astonishing frequency. These non-AUG initiation events are not simply errors but instead are used to generate or regulate proteins with key cellular functions; for example, during development or stress. Misregulation of non-AUG initiation events contributes to multiple human diseases, including cancer and neurodegeneration, and modulation of non-AUG usage may represent a novel therapeutic strategy. It is thus becoming increasingly clear that start codon selection is regulated by many trans-acting initiation factors as well as sequence/structural elements within messenger RNAs and that non-AUG translation has a profound impact on cellular states.
Keywords: RAN translation; eIF2A; eIF2D; near-cognate; start codon; translation initiation.
© 2017 Kearse and Wilusz; Published by Cold Spring Harbor Laboratory Press.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
