Dense Bicoid hubs accentuate binding along the morphogen gradient
- PMID: 28982761
- PMCID: PMC5666676
- DOI: 10.1101/gad.305078.117
Dense Bicoid hubs accentuate binding along the morphogen gradient
Abstract
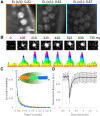
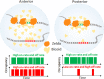
Morphogen gradients direct the spatial patterning of developing embryos; however, the mechanisms by which these gradients are interpreted remain elusive. Here we used lattice light-sheet microscopy to perform in vivo single-molecule imaging in early Drosophila melanogaster embryos of the transcription factor Bicoid that forms a gradient and initiates patterning along the anteroposterior axis. In contrast to canonical models, we observed that Bicoid binds to DNA with a rapid off rate throughout the embryo such that its average occupancy at target loci is on-rate-dependent. We further observed Bicoid forming transient "hubs" of locally high density that facilitate binding as factor levels drop, including in the posterior, where we observed Bicoid binding despite vanishingly low protein levels. We propose that localized modulation of transcription factor on rates via clustering provides a general mechanism to facilitate binding to low-affinity targets and that this may be a prevalent feature of other developmental transcription factors.
Keywords: Bicoid, Zelda; Drosophila; morphogen; single-molecule fluorescence; transcription factor dynamics.
© 2017 Mir et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
