Role of HIV-specific CD8+ T cells in pediatric HIV cure strategies after widespread early viral escape
- PMID: 28983013
- PMCID: PMC5679167
- DOI: 10.1084/jem.20162123
Role of HIV-specific CD8+ T cells in pediatric HIV cure strategies after widespread early viral escape
Abstract
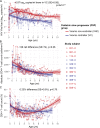
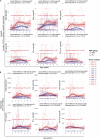
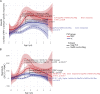
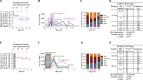
Recent studies have suggested greater HIV cure potential among infected children than adults. A major obstacle to HIV eradication in adults is that the viral reservoir is largely comprised of HIV-specific cytotoxic T lymphocyte (CTL) escape variants. We here evaluate the potential for CTL in HIV-infected slow-progressor children to play an effective role in "shock-and-kill" cure strategies. Two distinct subgroups of children were identified on the basis of viral load. Unexpectedly, in both groups, as in adults, HIV-specific CTL drove the selection of escape variants across a range of epitopes within the first weeks of infection. However, in HIV-infected children, but not adults, de novo autologous variant-specific CTL responses were generated, enabling the pediatric immune system to "corner" the virus. Thus, even when escape variants are selected in early infection, the capacity in children to generate variant-specific anti-HIV CTL responses maintains the potential for CTL to contribute to effective shock-and-kill cure strategies in pediatric HIV infection.
© 2017 Leitman et al.
Figures



References
-
- Addo M.M., Yu X.G., Rathod A., Cohen D., Eldridge R.L., Strick D., Johnston M.N., Corcoran C., Wurcel A.G., Fitzpatrick C.A., et al. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 77:2081–2092. 10.1128/JVI.77.3.2081-2092.2003 - DOI - PMC - PubMed
-
- Adland E., Paioni P., Thobakgale C., Laker L., Mori L., Muenchhoff M., Csala A., Clapson M., Flynn J., Novelli V., et al. 2015. Discordant Impact of HLA on Viral Replicative Capacity and Disease Progression in Pediatric and Adult HIV Infection. PLoS Pathog. 11:e1004954 10.1371/journal.ppat.1004954 - DOI - PMC - PubMed
-
- Allen T.M., Yu X.G., Kalife E.T., Reyor L.L., Lichterfeld M., John M., Cheng M., Allgaier R.L., Mui S., Frahm N., et al. 2005. De novo generation of escape variant-specific CD8+ T-cell responses following cytotoxic T-lymphocyte escape in chronic human immunodeficiency virus type 1 infection. J. Virol. 79:12952–12960. 10.1128/JVI.79.20.12952-12960.2005 - DOI - PMC - PubMed
-
- Almeida J.R., Price D.A., Papagno L., Arkoub Z.A., Sauce D., Bornstein E., Asher T.E., Samri A., Schnuriger A., Theodorou I., et al. 2007. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J. Exp. Med. 204:2473–2485. 10.1084/jem.20070784 - DOI - PMC - PubMed
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

