The GBAP1 pseudogene acts as a ceRNA for the glucocerebrosidase gene GBA by sponging miR-22-3p
- PMID: 28983119
- PMCID: PMC5629250
- DOI: 10.1038/s41598-017-12973-5
The GBAP1 pseudogene acts as a ceRNA for the glucocerebrosidase gene GBA by sponging miR-22-3p
Abstract
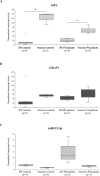
Mutations in the GBA gene, encoding lysosomal glucocerebrosidase, represent the major predisposing factor for Parkinson's disease (PD), and modulation of the glucocerebrosidase activity is an emerging PD therapy. However, little is known about mechanisms regulating GBA expression. We explored the existence of a regulatory network involving GBA, its expressed pseudogene GBAP1, and microRNAs. The high level of sequence identity between GBA and GBAP1 makes the pseudogene a promising competing-endogenous RNA (ceRNA), functioning as a microRNA sponge. After selecting microRNAs potentially targeting both transcripts, we demonstrated that miR-22-3p binds to and down-regulates GBA and GBAP1, and decreases their endogenous mRNA levels up to 70%. Moreover, over-expression of GBAP1 3'-untranslated region was able to sequester miR-22-3p, thus increasing GBA mRNA and glucocerebrosidase levels. The characterization of GBAP1 splicing identified multiple out-of-frame isoforms down-regulated by the nonsense-mediated mRNA decay, suggesting that GBAP1 levels and, accordingly, its ceRNA effect, are significantly modulated by this degradation process. Using skin-derived induced pluripotent stem cells of PD patients with GBA mutations and controls, we observed a significant GBA up-regulation during dopaminergic differentiation, paralleled by down-regulation of miR-22-3p. Our results describe the first microRNA controlling GBA and suggest that the GBAP1 non-coding RNA functions as a GBA ceRNA.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
A genetic variation in the CpG island of pseudogene GBAP1 promoter is associated with gastric cancer susceptibility.Cancer. 2019 Jul 15;125(14):2465-2473. doi: 10.1002/cncr.32081. Epub 2019 Apr 5. Cancer. 2019. PMID: 30951202
-
Redefining GBA gene structure unveils the ability of Cap-independent, IRES-dependent gene regulation.Commun Biol. 2022 Jul 13;5(1):639. doi: 10.1038/s42003-022-03577-5. Commun Biol. 2022. PMID: 35831491 Free PMC article.
-
Advances in GBA-associated Parkinson's disease--Pathology, presentation and therapies.Neurochem Int. 2016 Feb;93:6-25. doi: 10.1016/j.neuint.2015.12.004. Epub 2015 Dec 30. Neurochem Int. 2016. PMID: 26743617 Review.
-
Glucocerebrosidase activity in Parkinson's disease with and without GBA mutations.Brain. 2015 Sep;138(Pt 9):2648-58. doi: 10.1093/brain/awv179. Epub 2015 Jun 27. Brain. 2015. PMID: 26117366 Free PMC article.
-
Insights into GBA Parkinson's disease pathology and therapy with induced pluripotent stem cell model systems.Neurobiol Dis. 2019 Jul;127:1-12. doi: 10.1016/j.nbd.2019.01.023. Epub 2019 Jan 31. Neurobiol Dis. 2019. PMID: 30711484 Review.
Cited by
-
Epigenetic Analysis in Human Neurons: Considerations for Disease Modeling in PD.Front Neurosci. 2019 Apr 5;13:276. doi: 10.3389/fnins.2019.00276. eCollection 2019. Front Neurosci. 2019. PMID: 31024227 Free PMC article.
-
A compendious summary of Parkinson's disease patient-derived iPSCs in the first decade.Ann Transl Med. 2019 Nov;7(22):685. doi: 10.21037/atm.2019.11.16. Ann Transl Med. 2019. PMID: 31930086 Free PMC article. Review.
-
Meta-analysis of the Prognostic Value of microRNA-22 in Leukemia Patients.Technol Cancer Res Treat. 2023 Jan-Dec;22:15330338231212309. doi: 10.1177/15330338231212309. Technol Cancer Res Treat. 2023. PMID: 37942522 Free PMC article.
-
MiRNA Expression in Patients with Gaucher Disease Treated with Enzyme Replacement Therapy.Life (Basel). 2020 Dec 22;11(1):2. doi: 10.3390/life11010002. Life (Basel). 2020. PMID: 33375048 Free PMC article.
-
Whole-transcriptome sequencing in neural and non-neural tissues of a mouse model identifies miR-34a as a key regulator in SMA pathogenesis.Mol Ther Nucleic Acids. 2025 Feb 20;36(2):102490. doi: 10.1016/j.omtn.2025.102490. eCollection 2025 Jun 10. Mol Ther Nucleic Acids. 2025. PMID: 40125274 Free PMC article.
References
-
- De Fost M, Aerts JM, Hollak CE. Gaucher disease: from fundamental research to effective therapeutic interventions. Neth. J. Med. 2003;61:3–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

