Total rRNA-Seq Analysis Gives Insight into Bacterial, Fungal, Protozoal and Archaeal Communities in the Rumen Using an Optimized RNA Isolation Method
- PMID: 28983291
- PMCID: PMC5613150
- DOI: 10.3389/fmicb.2017.01814
Total rRNA-Seq Analysis Gives Insight into Bacterial, Fungal, Protozoal and Archaeal Communities in the Rumen Using an Optimized RNA Isolation Method
Abstract
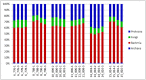
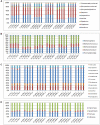
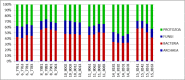
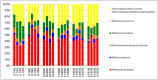
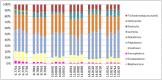
Advances in high throughput, next generation sequencing technologies have allowed an in-depth examination of biological environments and phenomena, and are particularly useful for culture-independent microbial community studies. Recently the use of RNA for metatranscriptomic studies has been used to elucidate the role of active microbes in the environment. Extraction of RNA of appropriate quality is critical in these experiments and TRIzol reagent is often used for maintaining stability of RNA molecules during extraction. However, for studies using rumen content there is no consensus on (1) the amount of rumen digesta to use or (2) the amount of TRIzol reagent to be used in RNA extraction procedures. This study evaluated the effect of using various quantities of ground rumen digesta and of TRIzol reagent on the yield and quality of extracted RNA. It also investigated the possibility of using lower masses of solid-phase rumen digesta and lower amounts of TRIzol reagent than is used currently, for extraction of RNA for metatranscriptomic studies. We found that high quality RNA could be isolated from 2 g of ground rumen digesta sample, whilst using 0.6 g of ground matter for RNA extraction and using 3 mL (a 5:1 TRIzol : extraction mass ratio) of TRIzol reagent. This represents a significant savings in the cost of RNA isolation. These lower masses and volumes were then applied in the RNA-Seq analysis of solid-phase rumen samples obtained from 6 Angus X Hereford beef heifers which had been fed a high forage diet (comprised of barley straw in a forage-to-concentrate ratio of 70:30) for 102 days. A bioinformatics analysis pipeline was developed in-house that generated relative abundance values of archaea, protozoa, fungi and bacteria in the rumen and also allowed the extraction of individual rRNA variable regions that could be analyzed in downstream molecular ecology programs. The average relative abundances of rRNA transcripts of archaea, bacteria, protozoa and fungi in our samples were 1.4 ± 0.06, 44.16 ± 1.55, 35.38 ± 1.64, and 16.37 ± 0.65% respectively. This represents the first study to define the relative active contributions of these populations to the rumen ecosystem and is especially important in defining the role of the anaerobic fungi and protozoa.
Keywords: RNA isolation; bioinformatics; metatranscriptomics; microbial diversity; rRNA-Seq.
Figures


References
-
- Andrews S. (2010). FastQC A Quality Control Tool for High Throughput Sequence Data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
LinkOut - more resources
Full Text Sources
Other Literature Sources

