Mobile Genetic Elements and Evolution of CRISPR-Cas Systems: All the Way There and Back
- PMID: 28985291
- PMCID: PMC5737515
- DOI: 10.1093/gbe/evx192
Mobile Genetic Elements and Evolution of CRISPR-Cas Systems: All the Way There and Back
Abstract
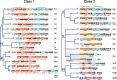
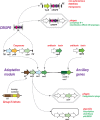
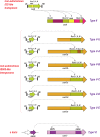
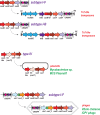
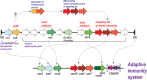
The Clustered Regularly Interspaced Palindromic Repeats (CRISPR)-CRISPR-associated proteins (Cas) systems of bacterial and archaeal adaptive immunity show multifaceted evolutionary relationships with at least five classes of mobile genetic elements (MGE). First, the adaptation module of CRISPR-Cas that is responsible for the formation of the immune memory apparently evolved from a Casposon, a self-synthesizing transposon that employs the Cas1 protein as the integrase and might have brought additional cas genes to the emerging immunity loci. Second, a large subset of type III CRISPR-Cas systems recruited a reverse transcriptase from a Group II intron, providing for spacer acquisition from RNA. Third, effector nucleases of Class 2 CRISPR-Cas systems that are responsible for the recognition and cleavage of the target DNA were derived from transposon-encoded TnpB nucleases, most likely, on several independent occasions. Fourth, accessory nucleases in some variants of types I and III toxin and type VI effectors RNases appear to be ultimately derived from toxin nucleases of microbial toxin-antitoxin modules. Fifth, the opposite direction of evolution is manifested in the recruitment of CRISPR-Cas systems by a distinct family of Tn7-like transposons that probably exploit the capacity of CRISPR-Cas to recognize unique DNA sites to facilitate transposition as well as by bacteriophages that employ them to cope with host defense. Additionally, individual Cas proteins, such as the Cas4 nuclease, were recruited by bacteriophages and transposons. The two-sided evolutionary connection between CRISPR-Cas and MGE fits the "guns for hire" paradigm whereby homologous enzymatic machineries, in particular nucleases, are shuttled between MGE and defense systems and are used alternately as means of offense or defense.
Keywords: CRISPR adaptation; CRISPR effector modules; CRISPR-Cas systems; casposons; mobile genetic elements.
Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution 2017. This work is written by US Government employees and is in the public domain in the US.
Figures
References
-
- Allen SE, Nowacki M.. 2017. Necessity is the mother of invention: ciliates, transposons, and transgenerational inheritance. Trends Genet. 33(3):197–207. - PubMed
-
- Amitai G, Sorek R.. 2016. CRISPR-Cas adaptation: insights into the mechanism of action. Nat Rev Microbiol. 14(2):67–76. - PubMed
-
- Aravin AA, Hannon GJ, Brennecke J.. 2007. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318(5851):761–764. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

