The Revolution Continues: Newly Discovered Systems Expand the CRISPR-Cas Toolkit
- PMID: 28985502
- PMCID: PMC5683099
- DOI: 10.1016/j.molcel.2017.09.007
The Revolution Continues: Newly Discovered Systems Expand the CRISPR-Cas Toolkit
Abstract
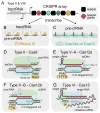
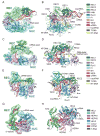
CRISPR-Cas systems defend prokaryotes against bacteriophages and mobile genetic elements and serve as the basis for revolutionary tools for genetic engineering. Class 2 CRISPR-Cas systems use single Cas endonucleases paired with guide RNAs to cleave complementary nucleic acid targets, enabling programmable sequence-specific targeting with minimal machinery. Recent discoveries of previously unidentified CRISPR-Cas systems have uncovered a deep reservoir of potential biotechnological tools beyond the well-characterized Type II Cas9 systems. Here we review the current mechanistic understanding of newly discovered single-protein Cas endonucleases. Comparison of these Cas effectors reveals substantial mechanistic diversity, underscoring the phylogenetic divergence of related CRISPR-Cas systems. This diversity has enabled further expansion of CRISPR-Cas biotechnological toolkits, with wide-ranging applications from genome editing to diagnostic tools based on various Cas endonuclease activities. These advances highlight the exciting prospects for future tools based on the continually expanding set of CRISPR-Cas systems.
Keywords: C2c1; C2c2; CRISPR-Cas; Cas12; Cas13; Cas9; Cpf1; RNA-guided DNA cleavage; RNA-guided RNA cleavage; genome editing.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures



References
-
- Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DBT, Shmakov S, Makarova KS, Semenova E, Minakhin L, Severinov K, Regev A, Lander ES, Koonin EV, Zhang F. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016:353. doi: 10.1126/science.aaf5573. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

