Chromatin and Single-Cell RNA-Seq Profiling Reveal Dynamic Signaling and Metabolic Transitions during Human Spermatogonial Stem Cell Development
- PMID: 28985528
- PMCID: PMC5832720
- DOI: 10.1016/j.stem.2017.09.003
Chromatin and Single-Cell RNA-Seq Profiling Reveal Dynamic Signaling and Metabolic Transitions during Human Spermatogonial Stem Cell Development
Abstract
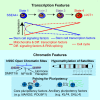
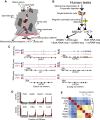
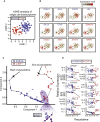
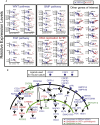
Human adult spermatogonial stem cells (hSSCs) must balance self-renewal and differentiation. To understand how this is achieved, we profiled DNA methylation and open chromatin (ATAC-seq) in SSEA4+ hSSCs, analyzed bulk and single-cell RNA transcriptomes (RNA-seq) in SSEA4+ hSSCs and differentiating c-KIT+ spermatogonia, and performed validation studies via immunofluorescence. First, DNA hypomethylation at embryonic developmental genes supports their epigenetic "poising" in hSSCs for future/embryonic expression, while core pluripotency genes (OCT4 and NANOG) were transcriptionally and epigenetically repressed. Interestingly, open chromatin in hSSCs was strikingly enriched in binding sites for pioneer factors (NFYA/B, DMRT1, and hormone receptors). Remarkably, single-cell RNA-seq clustering analysis identified four cellular/developmental states during hSSC differentiation, involving major transitions in cell-cycle and transcriptional regulators, splicing and signaling factors, and glucose/mitochondria regulators. Overall, our results outline the dynamic chromatin/transcription landscape operating in hSSCs and identify crucial molecular pathways that accompany the transition from quiescence to proliferation and differentiation.
Keywords: DNA methylation; hormone receptors; human spermatogonial stem cells; metabolism; open chromatin; pluripotency; single-cell RNA-seq; spermatogenesis.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures



Comment in
-
Epigenomic and single-cell profiling of human spermatogonial stem cells.Stem Cell Investig. 2018 Apr 24;5:11. doi: 10.21037/sci.2018.04.04. eCollection 2018. Stem Cell Investig. 2018. PMID: 29782571 Free PMC article. No abstract available.
References
-
- Boitani C., Di Persio S., Esposito V., Vicini E. Spermatogonial cells: Mouse, monkey and man comparison. Semin. Cell Dev. Biol. 2016;59:79–88. - PubMed
-
- Erkek S., Hisano M., Liang C.-Y., Gill M., Murr R., Dieker J., Schübeler D., van der Vlag J., Stadler M.B., Peters A.H.F.M. Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nat. Struct. Mol. Biol. 2013;20:868–875. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

