Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma
- PMID: 28985567
- PMCID: PMC5659841
- DOI: 10.1016/j.cell.2017.09.027
Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma
Abstract
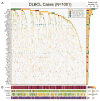
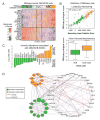
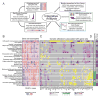
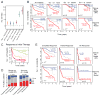
Diffuse large B cell lymphoma (DLBCL) is the most common form of blood cancer and is characterized by a striking degree of genetic and clinical heterogeneity. This heterogeneity poses a major barrier to understanding the genetic basis of the disease and its response to therapy. Here, we performed an integrative analysis of whole-exome sequencing and transcriptome sequencing in a cohort of 1,001 DLBCL patients to comprehensively define the landscape of 150 genetic drivers of the disease. We characterized the functional impact of these genes using an unbiased CRISPR screen of DLBCL cell lines to define oncogenes that promote cell growth. A prognostic model comprising these genetic alterations outperformed current established methods: cell of origin, the International Prognostic Index comprising clinical variables, and dual MYC and BCL2 expression. These results comprehensively define the genetic drivers and their functional roles in DLBCL to identify new therapeutic opportunities in the disease.
Keywords: DLBCL; TCGA; The Cancer Genome Atlas; diffuse large B cell lymphoma; exome sequencing; genetic mutations.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Untangling the Web of Lymphoma Somatic Mutations.Cell. 2017 Oct 5;171(2):270-272. doi: 10.1016/j.cell.2017.09.031. Cell. 2017. PMID: 28985559
References
-
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. - PubMed
-
- Andrews S. FastQC: a quality control tool for high throughput sequence data 2010
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

