Mammalian APE1 controls miRNA processing and its interactome is linked to cancer RNA metabolism
- PMID: 28986522
- PMCID: PMC5630600
- DOI: 10.1038/s41467-017-00842-8
Mammalian APE1 controls miRNA processing and its interactome is linked to cancer RNA metabolism
Abstract
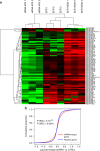
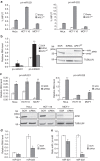
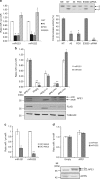
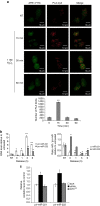
Mammalian apurinic/apyrimidinic endonuclease 1 is a DNA repair enzyme involved in genome stability and expression of genes involved in oxidative stress responses, tumor progression and chemoresistance. However, the molecular mechanisms underlying the role of apurinic/apyrimidinic endonuclease 1 in these processes are still unclear. Recent findings point to a novel role of apurinic/apyrimidinic endonuclease 1 in RNA metabolism. Through the characterization of the interactomes of apurinic/apyrimidinic endonuclease 1 with RNA and other proteins, we demonstrate here a role for apurinic/apyrimidinic endonuclease 1 in pri-miRNA processing and stability via association with the DROSHA-processing complex during genotoxic stress. We also show that endonuclease activity of apurinic/apyrimidinic endonuclease 1 is required for the processing of miR-221/222 in regulating expression of the tumor suppressor PTEN. Analysis of a cohort of different cancers supports the relevance of our findings for tumor biology. We also show that apurinic/apyrimidinic endonuclease 1 participates in RNA-interactomes and protein-interactomes involved in cancer development, thus indicating an unsuspected post-transcriptional effect on cancer genes.APE1 plays an important role in the cellular response to oxidative stress, and mutations are linked to tumor progression and chemoresistance. Here, the authors characterize the interactions of APE1 with RNA and demonstrate a role in microRNA processing.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
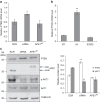


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

