Selective Cell-Surface Expression of Triheteromeric NMDA Receptors
- PMID: 28986871
- PMCID: PMC7286135
- DOI: 10.1007/978-1-4939-7321-7_7
Selective Cell-Surface Expression of Triheteromeric NMDA Receptors
Abstract
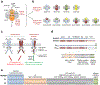
The NMDA-type ionotropic glutamate receptors play pivotal roles in many brain functions, but are also involved in numerous brain disorders. Seven NMDA receptor subunits exist (GluN1, GluN2A-D, and GluN3A-B) that assemble into a diverse array of tetrameric receptor subtypes with distinct functional properties and physiological roles. Most NMDA receptors are composed of two GluN1 and two GluN2 subunits, which can assemble into four diheteromeric receptor subtypes composed of GluN1 and one type of GluN2 subunit (e.g., GluN1/2A), and presumably also six triheteromeric receptor subtypes composed of GluN1 and two different GluN2 subunits (e.g., GluN1/2A/2B). Despite accumulating evidence that a large proportion of native NMDA receptors are triheteromers, little is known about their function and pharmacology due to the lack of methods to faithfully express triheteromeric NMDA receptors in heterologous expression systems. The problem is that co-expression of GluN1 with two different GluN2 subunits generates two distinct diheteromeric receptor subtypes as well as one triheteromeric receptor subtype, thereby confounding studies on a homogenous population of triheteromeric NMDA receptors. Here, we will describe a method to selectively express recombinant triheteromeric GluN1/2A/2B receptors without interfering co-expression of diheteromeric GluN1/2A and GluN1/2B receptors. This method enables quantitative evaluation of functional and pharmacological properties of triheteromeric GluN1/2A/2B receptors, which are presumably the most abundant NMDA receptors in the adult cortex and hippocampus.
Keywords: Assembly; Coiled-coil; Endoplasmic reticulum; Ionotropic glutamate receptor; Ligand-gated ion channel; Retention signals; Trafficking; Xenopus oocytes.
Figures
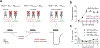
References
-
- Perez-Otano I, Larsen RS, Wesseling JF (2016) Emerging roles of GluN3-containing NMDA receptors in the CNS. Nat Rev Neurosci 17 (10):623–635 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

