Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer
- PMID: 28988769
- PMCID: PMC5687509
- DOI: 10.1016/j.cell.2017.09.007
Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer
Erratum in
-
Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer.Cell. 2018 Aug 9;174(4):1033. doi: 10.1016/j.cell.2018.07.036. Cell. 2018. PMID: 30096301 Free PMC article. No abstract available.
Abstract
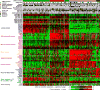
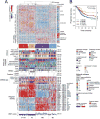
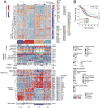
We report a comprehensive analysis of 412 muscle-invasive bladder cancers characterized by multiple TCGA analytical platforms. Fifty-eight genes were significantly mutated, and the overall mutational load was associated with APOBEC-signature mutagenesis. Clustering by mutation signature identified a high-mutation subset with 75% 5-year survival. mRNA expression clustering refined prior clustering analyses and identified a poor-survival "neuronal" subtype in which the majority of tumors lacked small cell or neuroendocrine histology. Clustering by mRNA, long non-coding RNA (lncRNA), and miRNA expression converged to identify subsets with differential epithelial-mesenchymal transition status, carcinoma in situ scores, histologic features, and survival. Our analyses identified 5 expression subtypes that may stratify response to different treatments.
Keywords: APOBEC mutation; DNA methylation; basal mRNA subtype; lncRNA transcriptome; luminal mRNA subtype; microRNA; muscle-invasive bladder cancer; neoantigen; neuronal subtype; regulon.
Copyright © 2017 Elsevier Inc. All rights reserved.
Conflict of interest statement
S.P.L. has received investigator initiated research funding from Endo Pharmaceuticals; support for a clinical trial from FKD and Viventia; is a consultant for UroGen, Vaxiion, Nucleix and BioCancell. A.D.C., G.G. and M.M have received research funding from Bayer AG. E.A.G. is now employed by GenomeDx Biosciences. M.M. has no current conflicts; was previously an equity holder in and consultant for Foundation Medicine. G.B.M. is a member of the scientific advisory board and receives research support from AstraZeneca. D.J.M. has stock options in ApoCell, Inc. J.B. has a paid consultancy with Pfizer; has received advisory board and lecture fees from Merck; has received advisory board fees from Genentech; has given uncompensated presentations at Genentech. C.J.W. is a cofounder and advisory board member of Neon Therapeutics. No other conflicts of interest declared.
Figures



Comment in
-
Re: Comprehensive Molecular Characterization of Muscle-invasive Bladder Cancer.Eur Urol. 2018 May;73(5):808-809. doi: 10.1016/j.eururo.2017.12.010. Epub 2017 Dec 20. Eur Urol. 2018. PMID: 29274865 No abstract available.
References
MeSH terms
Substances
Grants and funding
- U24 CA143882/CA/NCI NIH HHS/United States
- HHSN261201000021C/CP/NCI NIH HHS/United States
- P50 CA100632/CA/NCI NIH HHS/United States
- U24 CA143866/CA/NCI NIH HHS/United States
- P30 CA016086/CA/NCI NIH HHS/United States
- U24 CA143840/CA/NCI NIH HHS/United States
- U24 CA143843/CA/NCI NIH HHS/United States
- U24 CA210974/CA/NCI NIH HHS/United States
- U24 CA143858/CA/NCI NIH HHS/United States
- U24 CA210949/CA/NCI NIH HHS/United States
- HHSN261200800001C/RC/CCR NIH HHS/United States
- P50 CA091846/CA/NCI NIH HHS/United States
- P30 CA016672/CA/NCI NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- U24 CA143835/CA/NCI NIH HHS/United States
- HHSN261201000032C/CA/NCI NIH HHS/United States
- U24 CA210950/CA/NCI NIH HHS/United States
- P01 CA120964/CA/NCI NIH HHS/United States
- U24 CA143845/CA/NCI NIH HHS/United States
- U24 CA143799/CA/NCI NIH HHS/United States
- U01 CA168394/CA/NCI NIH HHS/United States
- U01 CA217842/CA/NCI NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- U24 CA144025/CA/NCI NIH HHS/United States
- R50 CA211482/CA/NCI NIH HHS/United States
- R01 CA155010/CA/NCI NIH HHS/United States
- U24 CA143848/CA/NCI NIH HHS/United States
- U54 HG003079/HG/NHGRI NIH HHS/United States
- R01 CA178744/CA/NCI NIH HHS/United States
- U24 CA143883/CA/NCI NIH HHS/United States
- U24 CA210999/CA/NCI NIH HHS/United States
- U24 CA143867/CA/NCI NIH HHS/United States
- U24 CA199461/CA/NCI NIH HHS/United States
- U24 CA209851/CA/NCI NIH HHS/United States
- HHSN261200800001E/CA/NCI NIH HHS/United States
- R50 CA221675/CA/NCI NIH HHS/United States
- K23 CA160664/CA/NCI NIH HHS/United States
- UL1 TR000371/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

