Rapid determination of medulloblastoma subgroup affiliation with mass spectrometry using a handheld picosecond infrared laser desorption probe
- PMID: 28989676
- PMCID: PMC5628578
- DOI: 10.1039/c7sc01974b
Rapid determination of medulloblastoma subgroup affiliation with mass spectrometry using a handheld picosecond infrared laser desorption probe
Abstract
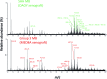
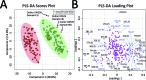
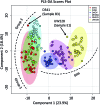
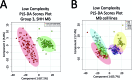
Medulloblastoma (MB), the most prevalent malignant childhood brain tumour, consists of at least 4 distinct subgroups each of which possesses a unique survival rate and response to treatment. To rapidly determine MB subgroup affiliation in a manner that would be actionable during surgery, we subjected murine xenograft tumours of two MB subgroups (SHH and Group 3) to Mass Spectrometry (MS) profiling using a handheld Picosecond InfraRed Laser (PIRL) desorption probe and interface developed by our group. This platform provides real time MS profiles of tissue based on laser desorbed lipids and small molecules with only 5-10 seconds of sampling. PIRL-MS analysis of ex vivo MB tumours offered a 98% success rate in subgroup determination, observed over 194 PIRL-MS datasets collected from 19 independent tumours (∼10 repetitions each) utilizing 6 different established MB cell lines. Robustness was verified by a 5%-leave-out-and-remodel test. PIRL ablated tissue material was collected on a filter paper and subjected to high resolution LC-MS to provide ion identity assignments for the m/z values that contribute most to the statistical discrimination between SHH and Group 3 MB. Based on this analysis, rapid classification of MB with PIRL-MS utilizes a variety of fatty acid chains, glycerophosphates, glycerophosphoglycerols and glycerophosphocholines rapidly extracted from the tumours. In this work, we provide evidence that 5-10 seconds of sampling from ex vivo MB tissue with PIRL-MS can allow robust tumour subgroup classification, and have identified several biomarker ions responsible for the statistical discrimination of MB Group 3 and the SHH subgroup. The existing PIRL-MS platform used herein offers capabilities for future in vivo use.
Figures
References
-
- Ramaswamy V., Remke M., Bouffet E., Bailey S., Clifford S. C., Doz F., Kool M., Dufour C., Vassal G., Milde T., Witt O., von Hoff K., Pietsch T., Northcott P. A., Gajjar A., Robinson G. W., Padovani L., Andre N., Massimino M., Pizer B., Packer R., Rutkowski S., Pfister S. M., Taylor M. D., Pomeroy S. L. Acta Neuropathol. 2016;131:821–831. - PMC - PubMed
-
- Gottardo N. G., Hansford J. R., McGlade J. P., Alvaro F., Ashley D. M., Bailey S., Baker D. L., Bourdeaut F., Cho Y. J., Clay M., Clifford S. C., Cohn R. J., Cole C. H., Dallas P. B., Downie P., Doz F., Ellison D. W., Endersby R., Fisher P. G., Hassall T., Heath J. A., Hii H. L., Jones D. T., Junckerstorff R., Kellie S., Kool M., Kotecha R. S., Lichter P., Laughton S. J., Lee S., McCowage G., Northcott P. A., Olson J. M., Packer R. J., Pfister S. M., Pietsch T., Pizer B., Pomeroy S. L., Remke M., Robinson G. W., Rutkowski S., Schoep T., Shelat A. A., Stewart C. F., Sullivan M., Taylor M. D., Wainwright B., Walwyn T., Weiss W. A., Williamson D., Gajjar A. Acta Neuropathol. 2014;127:189–201. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

