Structural Biology of Proline Catabolic Enzymes
- PMID: 28990412
- PMCID: PMC6338584
- DOI: 10.1089/ars.2017.7374
Structural Biology of Proline Catabolic Enzymes
Abstract
Significance: Proline catabolism refers to the 4-electron oxidation of proline to glutamate catalyzed by the enzymes proline dehydrogenase (PRODH) and l-glutamate γ-semialdehyde dehydrogenase (GSALDH, or ALDH4A1). These enzymes and the intermediate metabolites of the pathway have been implicated in tumor growth and suppression, metastasis, hyperprolinemia metabolic disorders, schizophrenia susceptibility, life span extension, and pathogen virulence and survival. In some bacteria, PRODH and GSALDH are combined into a bifunctional enzyme known as proline utilization A (PutA). PutAs are not only virulence factors in some pathogenic bacteria but also fascinating systems for studying the coordination of metabolic enzymes via substrate channeling. Recent Advances: The past decade has seen an explosion of structural data for proline catabolic enzymes. This review surveys these structures, emphasizing protein folds, substrate recognition, oligomerization, kinetic mechanisms, and substrate channeling in PutA.
Critical issues: Major unsolved structural targets include eukaryotic PRODH, the complex between monofunctional PRODH and monofunctional GSALDH, and the largest of all PutAs, trifunctional PutA. The structural basis of PutA-membrane association is poorly understood. Fundamental aspects of substrate channeling in PutA remain unknown, such as the identity of the channeled intermediate, how the tunnel system is activated, and the roles of ancillary tunnels.
Future directions: New approaches are needed to study the molecular and in vivo mechanisms of substrate channeling. With the discovery of the proline cycle driving tumor growth and metastasis, the development of inhibitors of proline metabolic enzymes has emerged as an exciting new direction. Structural biology will be important in these endeavors.
Keywords: aldehyde dehydrogenase 4A1; proline dehydrogenase; proline utilization A; protein oligomerization; substrate channeling.
Figures

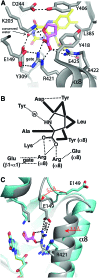










References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

