Redox Regulation of Heme Oxygenase-2 and the Transcription Factor, Rev-Erb, Through Heme Regulatory Motifs
- PMID: 28990415
- PMCID: PMC6217750
- DOI: 10.1089/ars.2017.7368
Redox Regulation of Heme Oxygenase-2 and the Transcription Factor, Rev-Erb, Through Heme Regulatory Motifs
Abstract
Significance: Heme binds to and serves as a cofactor for a myriad of proteins that are involved in diverse biological processes. Hemoproteins also exhibit varying modes of heme binding, suggesting that the protein environment contributes to the functional versatility of this prosthetic group. The subject of this review is a subset of hemoproteins that contain at least one heme regulatory motif (HRM), which is a short sequence containing a Cys-Pro core that, in many cases, binds heme with the Cys acting as an axial ligand. Recent Advances: As more details about HRM-containing proteins are uncovered, some underlying commonalities are emerging, including a role in regulating protein stability. Further, the cysteines of some HRMs have been shown to form disulfide bonds. Because the cysteines must be in the reduced, dithiol form to act as a heme axial ligand, heme binds at these sites in a redox-regulated manner, as demonstrated for heme oxygenase-2 (HO2) and Rev-erbβ.
Critical issues: HRM-containing proteins have wide variations in heme affinity, utilize different axial ligand schemes, and exhibit differences in the ability to act as a redox sensor-all while having a wide variety of biological functions. Here, we highlight HO2 and Rev-erbβ to illustrate the similarities and differences between two hemoproteins that contain HRMs acting as redox sensors.
Future directions: HRMs acting as redox sensors may be applicable to other HRM-containing proteins as many contain multiple HRMs and/or other cysteine residues, which may become more evident as the functional significance of HRMs is probed in additional proteins.
Keywords: disulfide; heme; heme oxygenase; heme regulatory motif; nuclear receptor; redox regulation.
Figures


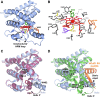


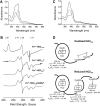
 ), and HO2solR (—) in 50 mM Tris (pH 8.0) and 50 mM KCl at 20°C. (B) EPR spectra of the Fe3+-heme-bound forms of HO2core, HO2solR, HO2tailR, and the His45Ala variant of HO2solR. The g values are indicated above the spectra. (C) Absorbance spectra of 5 μM Fe3+-heme-bound forms of HO2tailR (—), HO2tail(C282A)R (
), and HO2solR (—) in 50 mM Tris (pH 8.0) and 50 mM KCl at 20°C. (B) EPR spectra of the Fe3+-heme-bound forms of HO2core, HO2solR, HO2tailR, and the His45Ala variant of HO2solR. The g values are indicated above the spectra. (C) Absorbance spectra of 5 μM Fe3+-heme-bound forms of HO2tailR (—), HO2tail(C282A)R ( ), and HO2tail(C265A)R (…) in 50 mM Tris (pH 8.0) and 50 mM KCl at 20°C. (D) A model of heme binding to the oxidized form of HO2sol, which has a single heme-binding site due to the participation of Cys265 and Cys282 in a disulfide bond, and the reduced form of HO2sol, which has three heme-binding sites. Below each heme-binding site are the characteristics of that site, including the maximum absorbance, the g-values, the Kd values obtained by equilibrium titrations [Kd (equil)], and the koff/kon values obtained by kinetic measurements. The figures in (A–C) are reprinted (adapted) with permission from Fleischhacker et al. (20). Copyright (2015) American Chemical Society. EPR, electron paramagnetic resonance; HO2core, HO2 spanning residues 1-248; HO2R, HO2 in the disulfide bond-reduced form; HO2sol, HO2 spanning residues 1-288; HO2tail, HO2 spanning residues 213-288. EPR, electron paramagnetic resonance; HO2core, HO2 spanning residues 1-248; HO2R, HO2 in the disulfide bond-reduced form; HO2sol, HO2 spanning residues 1-288; HO2tail, HO2 spanning residues 213-288.
), and HO2tail(C265A)R (…) in 50 mM Tris (pH 8.0) and 50 mM KCl at 20°C. (D) A model of heme binding to the oxidized form of HO2sol, which has a single heme-binding site due to the participation of Cys265 and Cys282 in a disulfide bond, and the reduced form of HO2sol, which has three heme-binding sites. Below each heme-binding site are the characteristics of that site, including the maximum absorbance, the g-values, the Kd values obtained by equilibrium titrations [Kd (equil)], and the koff/kon values obtained by kinetic measurements. The figures in (A–C) are reprinted (adapted) with permission from Fleischhacker et al. (20). Copyright (2015) American Chemical Society. EPR, electron paramagnetic resonance; HO2core, HO2 spanning residues 1-248; HO2R, HO2 in the disulfide bond-reduced form; HO2sol, HO2 spanning residues 1-288; HO2tail, HO2 spanning residues 213-288. EPR, electron paramagnetic resonance; HO2core, HO2 spanning residues 1-248; HO2R, HO2 in the disulfide bond-reduced form; HO2sol, HO2 spanning residues 1-288; HO2tail, HO2 spanning residues 213-288.References
-
- Akashi M. and Takumi T. The orphan nuclear receptor RORα regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol 12: 441–448, 2005 - PubMed
-
- Bohle DS, Dodd EL, Pinter TBJ, and Stillman MJ. Soluble diamagnetic model for malaria pigment: coordination chemistry of gallium(III)protoporphyrin-IX. Inorg Chem 51: 10747–10761, 2012 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
