Hypoxia activates cadherin-22 synthesis via eIF4E2 to drive cancer cell migration, invasion and adhesion
- PMID: 28991229
- PMCID: PMC5770212
- DOI: 10.1038/onc.2017.372
Hypoxia activates cadherin-22 synthesis via eIF4E2 to drive cancer cell migration, invasion and adhesion
Abstract
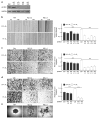
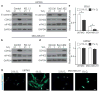
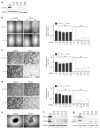
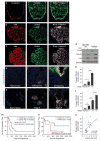
Hypoxia is a driver of cell movement in processes such as development and tumor progression. The cellular response to hypoxia involves a transcriptional program mediated by hypoxia-inducible factors, but translational control has emerged as a significant contributor. In this study, we demonstrate that a cell-cell adhesion molecule, cadherin-22, is upregulated in hypoxia via mTORC1-independent translational control by the initiation factor eIF4E2. We identify new functions of cadherin-22 as a hypoxia-specific cell-surface molecule involved in cancer cell migration, invasion and adhesion. Silencing eIF4E2 or cadherin-22 significantly impaired MDA-MB-231 breast carcinoma and U87MG glioblastoma cell migration and invasion only in hypoxia, while reintroduction of the respective exogenous gene restored the normal phenotype. Cadherin-22 was evenly distributed throughout spheroids and required for their formation and support of a hypoxic core. Conversely, E-cadherin translation was repressed by hypoxia and only expressed in the oxygenated cells of U87MG spheroids. Furthermore, immunofluorescence on paraffin-embedded human tissue from 40 glioma and 40 invasive ductal breast carcinoma patient specimens revealed that cadherin-22 expression colocalized with areas of hypoxia and significantly correlated with tumor grade and progression-free survival or stage and tumor size, respectively. This study broadens our understanding of tumor progression and metastasis by highlighting cadherin-22 as a potential new target of cancer therapy to disable hypoxic cancer cell motility and adhesion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Lin TA, Kong X, Haystead TA, Pause A, Belsham G, Sonenberg N, et al. PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science. 1994;266:653–656. - PubMed
-
- Rom E, Kim HC, Gingras AC, Marcotrigiano J, Favre D, Olsen H, et al. Cloning and characterization of 4EHP, a novel mammalian eIF4E-related cap-binding protein. J Biol Chem. 1998;273:13104–13109. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

