Stendomycin selectively inhibits TIM23-dependent mitochondrial protein import
- PMID: 28991239
- PMCID: PMC5945950
- DOI: 10.1038/nchembio.2493
Stendomycin selectively inhibits TIM23-dependent mitochondrial protein import
Abstract
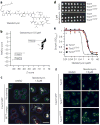
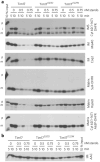
Tim17 and Tim23 are the main subunits of the TIM23 complex, one of the two major essential mitochondrial inner-membrane protein translocon machineries (TIMs). No chemical probes that specifically inhibit TIM23-dependent protein import were known to exist. Here we show that the natural product stendomycin, produced by Streptomyces hygroscopicus, is a potent and specific inhibitor of the TIM23 complex in yeast and mammalian cells. Furthermore, stendomycin-mediated blockage of the TIM23 complex does not alter normal processing of the major regulatory mitophagy kinase PINK1, but TIM23 is required to stabilize PINK1 on the outside of mitochondria to initiate mitophagy upon membrane depolarization.
Figures



Similar articles
-
Architecture of the TIM23 inner mitochondrial translocon and interactions with the matrix import motor.J Biol Chem. 2014 Oct 10;289(41):28689-96. doi: 10.1074/jbc.M114.588152. Epub 2014 Aug 25. J Biol Chem. 2014. PMID: 25157107 Free PMC article.
-
Tim50, a novel component of the TIM23 preprotein translocase of mitochondria.EMBO J. 2003 Feb 17;22(4):816-25. doi: 10.1093/emboj/cdg090. EMBO J. 2003. PMID: 12574118 Free PMC article.
-
Molecular pathway of mitochondrial preprotein import through the TOM-TIM23 supercomplex.Nat Struct Mol Biol. 2023 Dec;30(12):1996-2008. doi: 10.1038/s41594-023-01103-7. Epub 2023 Sep 11. Nat Struct Mol Biol. 2023. PMID: 37696957
-
Protein import into and across the mitochondrial inner membrane: role of the TIM23 and TIM22 translocons.Biochim Biophys Acta. 2002 Sep 2;1592(1):25-34. doi: 10.1016/s0167-4889(02)00261-6. Biochim Biophys Acta. 2002. PMID: 12191765 Review.
-
On the mechanism of preprotein import by the mitochondrial presequence translocase.Biochim Biophys Acta. 2010 Jun;1803(6):732-9. doi: 10.1016/j.bbamcr.2010.01.013. Epub 2010 Jan 25. Biochim Biophys Acta. 2010. PMID: 20100523 Review.
Cited by
-
Disease-Associated Genetic Variation in Human Mitochondrial Protein Import.Am J Hum Genet. 2019 May 2;104(5):784-801. doi: 10.1016/j.ajhg.2019.03.019. Am J Hum Genet. 2019. PMID: 31051112 Free PMC article. Review.
-
Structure-activity relationship of ipglycermide binding to phosphoglycerate mutases.J Biol Chem. 2021 Jan-Jun;296:100628. doi: 10.1016/j.jbc.2021.100628. Epub 2021 Apr 1. J Biol Chem. 2021. PMID: 33812994 Free PMC article.
-
PINK1 import regulation; a fine system to convey mitochondrial stress to the cytosol.BMC Biol. 2018 Jan 10;16(1):2. doi: 10.1186/s12915-017-0470-7. BMC Biol. 2018. PMID: 29325568 Free PMC article. Review.
-
Novel reporter of the PINK1-Parkin mitophagy pathway identifies its damage sensor in the import gate.bioRxiv [Preprint]. 2025 Feb 20:2025.02.19.639160. doi: 10.1101/2025.02.19.639160. bioRxiv. 2025. PMID: 40027798 Free PMC article. Preprint.
-
Visualization of Sirtuin 4 Distribution between Mitochondria and the Nucleus, Based on Bimolecular Fluorescence Self-Complementation.Cells. 2019 Dec 6;8(12):1583. doi: 10.3390/cells8121583. Cells. 2019. PMID: 31817668 Free PMC article.
References
-
- Aoun M, Tiranti V. Mitochondria: a crossroads for lipid metabolism defect in neurodegeneration with brain iron accumulation diseases. Int J Biochem Cell Biol. 2015;63:25–31. - PubMed
-
- Endo T, Yamamoto H, Esaki M. Functional cooperation and separation of translocators in protein import into mitochondria, the double-membrane bounded organelles. J Cell Sci. 2003;116:3259–3267. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

