Clonal dominance and transplantation dynamics in hematopoietic stem cell compartments
- PMID: 28991922
- PMCID: PMC5654265
- DOI: 10.1371/journal.pcbi.1005803
Clonal dominance and transplantation dynamics in hematopoietic stem cell compartments
Abstract
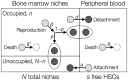
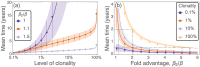
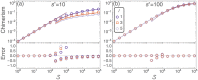
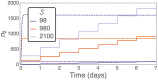
Hematopoietic stem cells in mammals are known to reside mostly in the bone marrow, but also transitively passage in small numbers in the blood. Experimental findings have suggested that they exist in a dynamic equilibrium, continuously migrating between these two compartments. Here we construct an individual-based mathematical model of this process, which is parametrised using existing empirical findings from mice. This approach allows us to quantify the amount of migration between the bone marrow niches and the peripheral blood. We use this model to investigate clonal hematopoiesis, which is a significant risk factor for hematologic cancers. We also analyse the engraftment of donor stem cells into non-conditioned and conditioned hosts, quantifying the impact of different treatment scenarios. The simplicity of the model permits a thorough mathematical analysis, providing deeper insights into the dynamics of both the model and of the real-world system. We predict the time taken for mutant clones to expand within a host, as well as chimerism levels that can be expected following transplantation therapy, and the probability that a preconditioned host is reconstituted by donor cells.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Kaushansky K, Lichtman MA, Prchal JT, Levi M, Press OW, Burns LJ, et al. Williams Hematology. 9th ed McGraw-Hill, New York; 2016.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

