Persulfidation proteome reveals the regulation of protein function by hydrogen sulfide in diverse biological processes in Arabidopsis
- PMID: 28992305
- PMCID: PMC5853657
- DOI: 10.1093/jxb/erx294
Persulfidation proteome reveals the regulation of protein function by hydrogen sulfide in diverse biological processes in Arabidopsis
Abstract
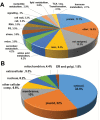
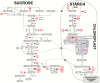
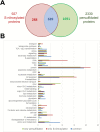
Hydrogen sulfide-mediated signaling pathways regulate many physiological and pathophysiological processes in mammalian and plant systems. The molecular mechanism by which hydrogen sulfide exerts its action involves the post-translational modification of cysteine residues to form a persulfidated thiol motif, a process called protein persulfidation. We have developed a comparative and quantitative proteomic analysis approach for the detection of endogenous persulfidated proteins in wild-type Arabidopsis and L-CYSTEINE DESULFHYDRASE 1 mutant leaves using the tag-switch method. The 2015 identified persulfidated proteins were isolated from plants grown under controlled conditions, and therefore, at least 5% of the entire Arabidopsis proteome may undergo persulfidation under baseline conditions. Bioinformatic analysis revealed that persulfidated cysteines participate in a wide range of biological functions, regulating important processes such as carbon metabolism, plant responses to abiotic and biotic stresses, plant growth and development, and RNA translation. Quantitative analysis in both genetic backgrounds reveals that protein persulfidation is mainly involved in primary metabolic pathways such as the tricarboxylic acid cycle, glycolysis, and the Calvin cycle, suggesting that this protein modification is a new regulatory component in these pathways.
Keywords: Cysteine; hydrogen sulfide; mass spectrometry; persulfidation; post-translational modification; proteomics.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures


References
-
- Álvarez C, Bermúdez MÁ, Romero LC, Gotor C, García I. 2012a. Cysteine homeostasis plays an essential role in plant immunity. New Phytologist 193, 165–177. - PubMed
-
- Álvarez C, García I, Romero LC, Gotor C. 2012c. Mitochondrial sulfide detoxification requires a functional isoform O-acetylserine(thiol)lyase C in Arabidopsis thaliana. Molecular Plant 5, 1217–1226. - PubMed
-
- Aroca A, Benito JM, Gotor C, Romero LC. PRIDE Archive, ProteomeXchange Consortium; 2017. https://www.ebi.ac.uk/pride/archive/projects/PXD005168 PXD006140https://www.ebi.ac.uk/pride/archive/projects/ Data from: Persulfidation proteome reveals the regulation of protein function by hydrogen sulfide in diverse biological processes in Arabidopsis.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

