Mechanisms of Tail-Anchored Membrane Protein Targeting and Insertion
- PMID: 28992441
- PMCID: PMC6343671
- DOI: 10.1146/annurev-cellbio-100616-060839
Mechanisms of Tail-Anchored Membrane Protein Targeting and Insertion
Abstract
Proper localization of membrane proteins is essential for the function of biological membranes and for the establishment of organelle identity within a cell. Molecular machineries that mediate membrane protein biogenesis need to not only achieve a high degree of efficiency and accuracy, but also prevent off-pathway aggregation events that can be detrimental to cells. The posttranslational targeting of tail-anchored proteins (TAs) provides tractable model systems to probe these fundamental issues. Recent advances in understanding TA-targeting pathways reveal sophisticated molecular machineries that drive and regulate these processes. These findings also suggest how an interconnected network of targeting factors, cochaperones, and quality control machineries together ensures robust membrane protein biogenesis.
Keywords: ATPase; chaperones; membrane protein biogenesis; protein quality control; protein targeting; tail-anchored protein.
Figures
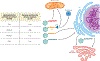


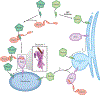
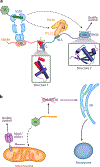
References
-
- Abell BM, Rabu C, Leznicki P, Young JC, High S. 2007. Post-translational integration of tail-anchored proteins is facilitated by defined molecular chaperones. J. Cell Sci. 120:1743–51 - PubMed
-
- Bae W, Lee YJ, Kim DH, Lee J, Kim S, et al. 2008. AKR2A-mediated import of chloroplast outer membrane proteins is essential for chloroplast biogenesis. Nat. Cell Biol. 10:220–27 - PubMed
-
- Beilharz T, Egan B, Silver PA, Hofmann K, Lithgow T. 2003. Bipartite signals mediate subcellular targeting of tail-anchored membrane proteins in Saccharomyces cerevisiae. J. Biol. Chem. 278:8219–23 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

