A functional analysis of TOEFAZ1 uncovers protein domains essential for cytokinesis in Trypanosoma brucei
- PMID: 28993462
- PMCID: PMC5702046
- DOI: 10.1242/jcs.207209
A functional analysis of TOEFAZ1 uncovers protein domains essential for cytokinesis in Trypanosoma brucei
Abstract
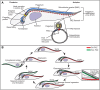
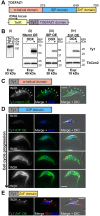
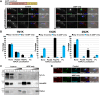
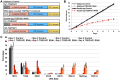
The parasite Trypanosoma brucei is highly polarized, including a flagellum that is attached along the cell surface by the flagellum attachment zone (FAZ). During cell division, the new FAZ positions the cleavage furrow, which ingresses from the anterior tip of the cell towards the posterior. We recently identified TOEFAZ1 (for 'Tip of the Extending FAZ protein 1') as an essential protein in trypanosome cytokinesis. Here, we analyzed the localization and function of TOEFAZ1 domains by performing overexpression and RNAi complementation experiments. TOEFAZ1 comprises three domains with separable functions: an N-terminal α-helical domain that may be involved in FAZ recruitment, a central intrinsically disordered domain that keeps the morphogenic kinase TbPLK at the new FAZ tip, and a C-terminal zinc finger domain necessary for TOEFAZ1 oligomerization. Both the N-terminal and C-terminal domains are essential for TOEFAZ1 function, but TbPLK retention at the FAZ is not necessary for cytokinesis. The feasibility of alternative cytokinetic pathways that do not employ TOEFAZ1 are also assessed. Our results show that TOEFAZ1 is a multimeric scaffold for recruiting proteins that control the timing and location of cleavage furrow ingression.
Keywords: Cleavage furrow ingression; Cytokinesis; Cytoskeleton; Flagellum attachment zone; Polo-like kinase; Trypanosoma brucei.
© 2017. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures




References
-
- Anderson W. A. and Ellis R.A. (1965). Ultrastructure of Trypanosoma lewisi: Flagellum, Microtubules, and the Kinetoplast. J. Protozool. Res. 12, 483-499. 10.1111/j.1550-7408.1965.tb03247.x - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

