A Ragulator-BORC interaction controls lysosome positioning in response to amino acid availability
- PMID: 28993468
- PMCID: PMC5716277
- DOI: 10.1083/jcb.201703094
A Ragulator-BORC interaction controls lysosome positioning in response to amino acid availability
Abstract
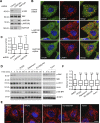
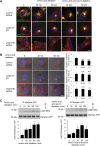
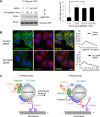
Lysosomes play key roles in the cellular response to amino acid availability. Depletion of amino acids from the medium turns off a signaling pathway involving the Ragulator complex and the Rag guanosine triphosphatases (GTPases), causing release of the inactive mammalian target of rapamycin complex 1 (mTORC1) serine/threonine kinase from the lysosomal membrane. Decreased phosphorylation of mTORC1 substrates inhibits protein synthesis while activating autophagy. Amino acid depletion also causes clustering of lysosomes in the juxtanuclear area of the cell, but the mechanisms responsible for this phenomenon are poorly understood. Herein we show that Ragulator directly interacts with BLOC-1-related complex (BORC), a multi-subunit complex previously found to promote lysosome dispersal through coupling to the small GTPase Arl8 and the kinesins KIF1B and KIF5B. Interaction with Ragulator exerts a negative regulatory effect on BORC that is independent of mTORC1 activity. Amino acid depletion strengthens this interaction, explaining the redistribution of lysosomes to the juxtanuclear area. These findings thus demonstrate that amino acid availability controls lysosome positioning through Ragulator-dependent, but mTORC1-independent, modulation of BORC.
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Figures

Comment in
-
Ragulator-a multifaceted regulator of lysosomal signaling and trafficking.J Cell Biol. 2017 Dec 4;216(12):3895-3898. doi: 10.1083/jcb.201710039. Epub 2017 Nov 14. J Cell Biol. 2017. PMID: 29138253 Free PMC article.
References
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

