Lamin B2 Modulates Nucleolar Morphology, Dynamics, and Function
- PMID: 28993479
- PMCID: PMC5705821
- DOI: 10.1128/MCB.00274-17
Lamin B2 Modulates Nucleolar Morphology, Dynamics, and Function
Abstract
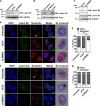
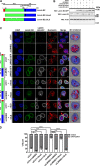
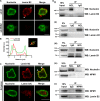
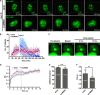
The nucleolus is required for ribosome biogenesis. Human cells have 2 or 3 nucleoli associated with nucleolar organizer region (NOR)-bearing chromosomes. An increase in number and altered nucleolar morphology define cancer cells. However, the mechanisms that modulate nucleolar morphology and function are unclear. Here we show that in addition to localizing at the nuclear envelope, lamin B2 localizes proximal to nucleolin at the granular component (GC) of the nucleolus and associates with the nucleolar proteins nucleolin and nucleophosmin. Lamin B2 knockdown severely disrupted the nucleolar morphology, which was rescued to intact and discrete nucleoli upon lamin B2 overexpression. Furthermore, two mutually exclusive lamin B2 deletion mutants, ΔHead and ΔSLS, rescued nuclear and nucleolar morphology defects, respectively, induced upon lamin B2 depletion, suggesting independent roles for lamin B2 at the nucleolus and nuclear envelope. Lamin B2 depletion increased nucleolin aggregation in the nucleoplasm, implicating lamin B2 in stabilizing nucleolin within the nucleolus. Lamin B2 knockdown upregulated nucleolus-specific 45S rRNA and upstream intergenic sequence (IGS) transcripts. The IGS transcripts colocalized with aggregates of nucleolin speckles, which were sustained in the nucleoplasm upon lamin B2 depletion. Taken together, these studies uncover a novel role for lamin B2 in modulating the morphology, dynamics, and function of the nucleolus.
Keywords: lamin; nucleolin; nucleolus; nucleophosmin; nucleus; rDNA; rRNA.
Copyright © 2017 Sen Gupta and Sengupta.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
