Precise, High-throughput Analysis of Bacterial Growth
- PMID: 28994811
- PMCID: PMC5752254
- DOI: 10.3791/56197
Precise, High-throughput Analysis of Bacterial Growth
Abstract
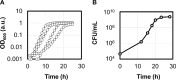
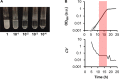
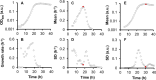
Bacterial growth is a central concept in the development of modern microbial physiology, as well as in the investigation of cellular dynamics at the systems level. Recent studies have reported correlations between bacterial growth and genome-wide events, such as genome reduction and transcriptome reorganization. Correctly analyzing bacterial growth is crucial for understanding the growth-dependent coordination of gene functions and cellular components. Accordingly, the precise quantitative evaluation of bacterial growth in a high-throughput manner is required. Emerging technological developments offer new experimental tools that allow updates of the methods used for studying bacterial growth. The protocol introduced here employs a microplate reader with a highly optimized experimental procedure for the reproducible and precise evaluation of bacterial growth. This protocol was used to evaluate the growth of several previously described Escherichia coli strains. The main steps of the protocol are as follows: the preparation of a large number of cell stocks in small vials for repeated tests with reproducible results, the use of 96-well plates for high-throughput growth evaluation, and the manual calculation of two major parameters (i.e., maximal growth rate and population density) representing the growth dynamics. In comparison to the traditional colony-forming unit (CFU) assay, which counts the cells that are cultured in glass tubes over time on agar plates, the present method is more efficient and provides more detailed temporal records of growth changes, but has a stricter detection limit at low population densities. In summary, the described method is advantageous for the precise and reproducible high-throughput analysis of bacterial growth, which can be used to draw conceptual conclusions or to make theoretical observations.
Similar articles
-
A method for high throughput determination of viable bacteria cell counts in 96-well plates.BMC Microbiol. 2012 Nov 13;12:259. doi: 10.1186/1471-2180-12-259. BMC Microbiol. 2012. PMID: 23148795 Free PMC article.
-
Colony-live--a high-throughput method for measuring microbial colony growth kinetics--reveals diverse growth effects of gene knockouts in Escherichia coli.BMC Microbiol. 2014 Jun 26;14:171. doi: 10.1186/1471-2180-14-171. BMC Microbiol. 2014. PMID: 24964927 Free PMC article.
-
Cultivating efficiency: high-throughput growth analysis of anaerobic bacteria in compact microplate readers.Microbiol Spectr. 2024 May 2;12(5):e0365023. doi: 10.1128/spectrum.03650-23. Epub 2024 Mar 19. Microbiol Spectr. 2024. PMID: 38501820 Free PMC article.
-
High-throughput system-wide engineering and screening for microbial biotechnology.Curr Opin Biotechnol. 2017 Aug;46:120-125. doi: 10.1016/j.copbio.2017.02.011. Epub 2017 Mar 24. Curr Opin Biotechnol. 2017. PMID: 28346890 Review.
-
Lessons from the organization of a proficiency testing program in food microbiology by interlaboratory comparison: analytical methods in use, impact of methods on bacterial counts and measurement uncertainty of bacterial counts.Food Microbiol. 2006 Feb;23(1):1-38. doi: 10.1016/j.fm.2005.01.010. Food Microbiol. 2006. PMID: 16942983 Review.
Cited by
-
Method for reproducible automated bacterial cell culture and measurement.Synth Biol (Oxf). 2022 Aug 10;7(1):ysac013. doi: 10.1093/synbio/ysac013. eCollection 2022. Synth Biol (Oxf). 2022. PMID: 36101862 Free PMC article.
-
Predicting the decision making chemicals used for bacterial growth.Sci Rep. 2019 May 10;9(1):7251. doi: 10.1038/s41598-019-43587-8. Sci Rep. 2019. PMID: 31076576 Free PMC article.
-
Small Molecule Inhibition of an Exopolysaccharide Modification Enzyme is a Viable Strategy To Block Pseudomonas aeruginosa Pel Biofilm Formation.Microbiol Spectr. 2023 Jun 15;11(3):e0029623. doi: 10.1128/spectrum.00296-23. Epub 2023 Apr 26. Microbiol Spectr. 2023. PMID: 37098898 Free PMC article.
-
Machine learning-assisted medium optimization revealed the discriminated strategies for improved production of the foreign and native metabolites.Comput Struct Biotechnol J. 2023 Apr 20;21:2654-2663. doi: 10.1016/j.csbj.2023.04.020. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37138901 Free PMC article.
-
Correlated chromosomal periodicities according to the growth rate and gene expression.Sci Rep. 2020 Sep 23;10(1):15531. doi: 10.1038/s41598-020-72389-6. Sci Rep. 2020. PMID: 32968121 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources