Clustergrammer, a web-based heatmap visualization and analysis tool for high-dimensional biological data
- PMID: 28994825
- PMCID: PMC5634325
- DOI: 10.1038/sdata.2017.151
Clustergrammer, a web-based heatmap visualization and analysis tool for high-dimensional biological data
Abstract
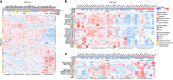
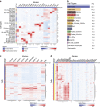
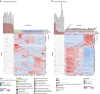
Most tools developed to visualize hierarchically clustered heatmaps generate static images. Clustergrammer is a web-based visualization tool with interactive features such as: zooming, panning, filtering, reordering, sharing, performing enrichment analysis, and providing dynamic gene annotations. Clustergrammer can be used to generate shareable interactive visualizations by uploading a data table to a web-site, or by embedding Clustergrammer in Jupyter Notebooks. The Clustergrammer core libraries can also be used as a toolkit by developers to generate visualizations within their own applications. Clustergrammer is demonstrated using gene expression data from the cancer cell line encyclopedia (CCLE), original post-translational modification data collected from lung cancer cells lines by a mass spectrometry approach, and original cytometry by time of flight (CyTOF) single-cell proteomics data from blood. Clustergrammer enables producing interactive web based visualizations for the analysis of diverse biological data.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
Data Citations
-
- Hornbeck P., Rikova K., Fernandez N., Ma’ayan A. 2017. figshare. https://doi.org/10.6084/m9.figshare.5339689 - DOI - PMC - PubMed
-
- Rahman A., Fernandez N., Ma’ayan A. 2017. figshare. https://doi.org/10.6084/m9.figshare.5339698 - DOI
-
- 2012. Gene Expression Omnibus. GSE36133
-
- Rouillard A., Fernandez N., Ma’ayan A. 2017. figshare. https://doi.org/10.6084/m9.figshare.5339707 - DOI
References
-
- Maaten L. V. D. & Hinton G. Visualizing data using t-SNE. Journal of Machine Learning Research 9, 2579–2605 (2008).
-
- Henson R. & Cetto L. The MATLAB bioinformatics toolbox. Encyclopedia of Genetics, Genomics, Proteomics and Bioinformatics 4, 105 (2005).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

