Age-related gene expression in luminal epithelial cells is driven by a microenvironment made from myoepithelial cells
- PMID: 29016359
- PMCID: PMC5680554
- DOI: 10.18632/aging.101298
Age-related gene expression in luminal epithelial cells is driven by a microenvironment made from myoepithelial cells
Abstract
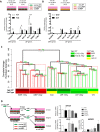
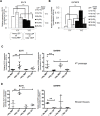
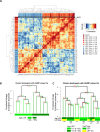
Luminal epithelial cells in the breast gradually alter gene and protein expression with age, appearing to lose lineage-specificity by acquiring myoepithelial-like characteristics. We hypothesize that the luminal lineage is particularly sensitive to microenvironment changes, and age-related microenvironment changes cause altered luminal cell phenotypes. To evaluate the effects of different microenvironments on the fidelity of epigenetically regulated luminal and myoepithelial gene expression, we generated a set of lineage-specific probes for genes that are controlled through DNA methylation. Culturing primary luminal cells under conditions that favor myoepithelial propogation led to their reprogramming at the level of gene methylation, and to a more myoepithelial-like expression profile. Primary luminal cells' lineage-specific gene expression could be maintained when they were cultured as bilayers with primary myoepithelial cells. Isogenic stromal fibroblast co-cultures were unable to maintain the luminal phenotype. Mixed-age luminal-myoepithelial bilayers revealed that luminal cells adopt transcription and methylation patterns consistent with the chronological age of the myoepithelial cells. We provide evidence that the luminal epithelial phenotype is exquisitely sensitive to microenvironment conditions, and that states of aging are cell non-autonomously communicated through microenvironment cues over at least one cell diameter.
Keywords: aging; breast cancer; epigenetic; mammary epithelia; microenvironment.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures




References
-
- Gudjonsson T, Adriance MC, Sternlicht MD, Petersen OW, Bissell MJ. Myoepithelial cells: their origin and function in breast morphogenesis and neoplasia. J Mammary Gland Biol Neoplasia. 2005;10:261–72. https://doi.org/10.1007/s10911-005-9586-4 - DOI - PMC - PubMed
-
- Howeedy AA, Virtanen I, Laitinen L, Gould NS, Koukoulis GK, Gould VE. Differential distribution of tenascin in the normal, hyperplastic, and neoplastic breast. Lab Invest. 1990;63:798–806. - PubMed
-
- Benz CC. Impact of aging on the biology of breast cancer. Crit Rev Oncol Hematol. 2008;66:65–74. https://doi.org/10.1016/j.critrevonc.2007.09.001 - DOI - PMC - PubMed
-
- Quong J, Eppenberger-Castori S, Moore D, 3rd, Scott GK, Birrer MJ, Kueng W, Eppenberger U, Benz CC. Age-dependent changes in breast cancer hormone receptors and oxidant stress markers. Breast Cancer Res Treat. 2002;76:221–36. https://doi.org/10.1023/A:1020886801674 - DOI - PubMed
-
- Pelissier FA, Garbe JC, Ananthanarayanan B, Miyano M, Lin C, Jokela T, Kumar S, Stampfer MR, Lorens JB, LaBarge MA. Age-related dysfunction in mechanotransduction impairs differentiation of human mammary epithelial progenitors. Cell Reports. 2014;7:1926–39. https://doi.org/10.1016/j.celrep.2014.05.021 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

