Broad Phylogenetic Occurrence of the Oxygen-Binding Hemerythrins in Bilaterians
- PMID: 29016798
- PMCID: PMC5629950
- DOI: 10.1093/gbe/evx181
Broad Phylogenetic Occurrence of the Oxygen-Binding Hemerythrins in Bilaterians
Abstract
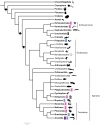
Animal tissues need to be properly oxygenated for carrying out catabolic respiration and, as such, natural selection has presumably favored special molecules that can reversibly bind and transport oxygen. Hemoglobins, hemocyanins, and hemerythrins (Hrs) fulfill this role, with Hrs being the least studied. Knowledge of oxygen-binding proteins is crucial for understanding animal physiology. Hr genes are present in the three domains of life, Archaea, Bacteria, and Eukaryota; however, within Animalia, Hrs has been reported only in marine species in six phyla (Annelida, Brachiopoda, Priapulida, Bryozoa, Cnidaria, and Arthropoda). Given this observed Hr distribution, whether all metazoan Hrs share a common origin is circumspect. We investigated Hr diversity and evolution in metazoans, by employing in silico approaches to survey for Hrs from of 120 metazoan transcriptomes and genomes. We found 58 candidate Hr genes actively transcribed in 36 species distributed in 11 animal phyla, with new records in Echinodermata, Hemichordata, Mollusca, Nemertea, Phoronida, and Platyhelminthes. Moreover, we found that "Hrs" reported from Cnidaria and Arthropoda were not consistent with that of other metazoan Hrs. Contrary to previous suggestions that Hr genes were absent in deuterostomes, we find Hr genes present in deuterostomes and were likely present in early bilaterians, but not in nonbilaterian animal lineages. As expected, the Hr gene tree did not mirror metazoan phylogeny, suggesting that Hrs evolutionary history was complex and besides the oxygen carrying capacity, the drivers of Hr evolution may also consist of secondary functional specializations of the proteins, like immunological functions.
Keywords: evolutionay history; metazoa; oxygen-binding protein; transcriptome.
© The Author(s) 2017. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures



References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3): 403–410. - PubMed
-
- Baert JL, Britel M, Sautière P, Malécha J.. 1992. Ovohemerythrin, a major 14-kDa yolk protein distinct from vitellogenin in leech. Eur J Biochem. 209(2): 563–569. - PubMed
-
- Brown CT, Howe A, Zhang Q, Pyrkosz AB, Brom TH.. 2012. A reference-free algorithm for computational normalization of shotgun sequencing data. arXiv:1203.4802
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

